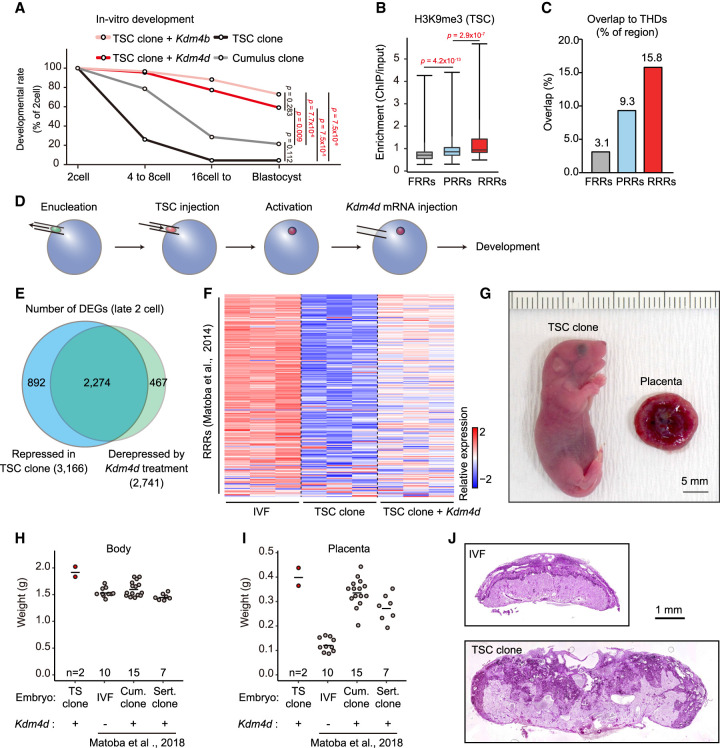
Figure 6.
H3K9me3 strongly impaired the developmental efficiency of TSC cloned embryos. (A) In vitro development of SCNT embryos derived from TSCs and cumulus cells. Injection of Kdm4d or Kdm4b mRNA greatly improved the blastocyst formation rate of TSC cloned embryos. P-values were calculated using Fisher's exact test. (B) Box plot showing H3K9me3 enrichment within FRRs, PRRs, and RRRs in TSCs. These regions are referred from Matoba et al. (2014). P-values were calculated using the Wilcoxon rank sum test. (C) Bar plot showing the percentage of overlapped regions between THDs and FRRs, PRRs, or RRRs. (D) Schematic illustration of SCNT procedure. Kdm4d mRNA was injected to erase H3K9me3 from the genome of TSC cloned embryos. (E) Venn diagram showing the overlap between the genes that failed to be activated in TSC cloned embryos and those derepressed in Kdm4d-treated TSC cloned embryos. Genes with Padj < 0.01 and fold change > 1.5 were classed as DEGs. (F) Heat map showing relative gene expression levels of the RRRs at the late two-cell stage. (G) A pup and a placenta derived by cloning from TSCs with Kdm4d mRNA injection. (H,I) Scatter plots showing the weight of the body (H) or the placenta (I) at birth (E19.5). Each dot represents a sample. Data for IVF-derived embryos, cumulus cell cloned embryos, and Sertoli cell cloned embryos are from Matoba et al. (2018). (J) Representative images of histological sections of term placentas stained with periodic acid Schiff (PAS). The TSC cloned placenta shows an expansion of the PAS-positive spongiotrophoblast layer and an irregular boundary with the PAS-negative labyrinthine layer.