Figure 3.
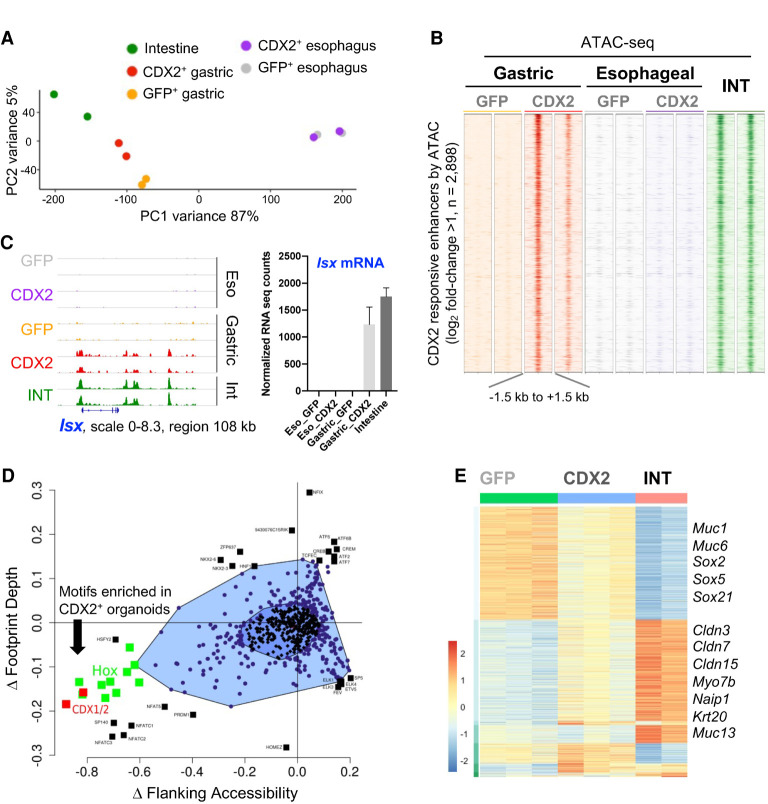
Intestinal TFs open intestinal enhancers in stomach but not esophageal organoids (A) Principal component analysis (PCA) of the 5000 most variable enhancers by ATAC-seq. Ectopic expression of CDX2 in stomach, but not in esophageal, organoids shifts the global enhancer profile toward that of intestinal epithelium. (B) ATAC-seq data at 2898 unique enhancers accessible in CDX2+ compared with GFP+ gastric organoids. These regions are closed in esophageal CDX2+ organoids. Each column shows an independent biological replicate, and all signals are quantile-normalized. (C) IGV data tracks of open chromatin at the intestine-restricted Isx locus in replicate samples. CDX2+ stomach organoids show increased access at multiple enhancers, in contrast to CDX2+ esophageal organoids, which lack these or other chromatin changes. Chromatin access was accompanied by gene activation (RNA-seq counts, mean ± SD) in gastric CDX2+ organoids. (D) Bagfoot analysis (Baek et al. 2017) of motif enrichment at sites that display metrics of TF binding; i.e., altered footprint depth or increased accessibility flanking ATAC peaks. CDX1/2 and HOX family motifs are significantly enriched in CDX2+ stomach organoids. (E) Hierarchical clustering of RNA-seq data from organoid lines (DESeq2, likelihood ratio test, q < 0.001) showing major groups of differentially expressed genes in GFP+ (n = 826) or intestinal (INT; n = 737) organoids. Exemplary genes within the major differential clusters are listed.