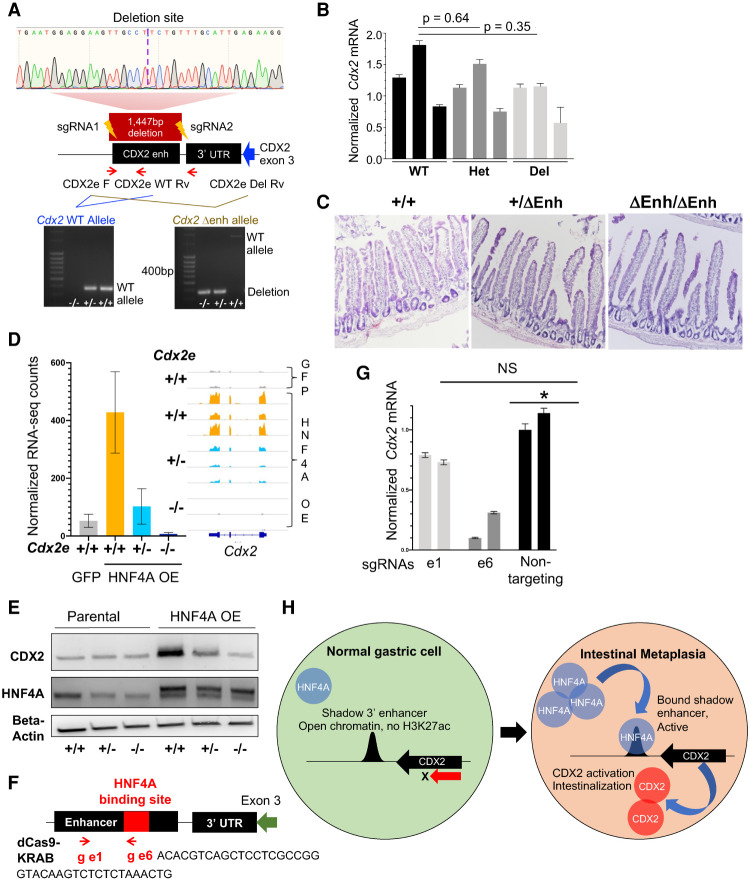
Figure 7.
HNF4A activates endogenous Cdx2 in gastric organoids through the shadow 3′ enhancer. (A) Design and validation of genetically modified mice with enhancer deletion by CRISPR–Cas9. Location of the enhancer downstream from the Cdx2 3′ UTR, targeting short guide (sg) RNAs, and primers used for PCR validation are shown; a maroon box delineates the 1447-bp enhancer deletion in the principal founder strain, confirmed by Sanger sequencing. (Bottom) Genotyping PCR for the enhancer-deleted (Δenh) strain. (Left) Differentiation of monoallelic from biallelic deletion (primers: CDX2e_F and CDX2e_WT_Rv). (Right) Enhancer deletion (primers: CDX2e_F and CDX2e_Del_Rv). (WT) Wild type. (B,C) Heterozygous and homozygous Δenh mice showed no loss of intestinal CDX2 by qRT-PCR (B) or changes in intestinal histology (C; representative images of duodenum) in three independent mice of each genotype. (D,E) HNF4A overexpression failed to induce CDX2 in gastric organoids cultured from enhancer-deleted homozygous mice, as shown by RNA-seq (D; confirmed by qRT-PCR) (Supplemental Fig. S7C) and immunoblots (E). CDX2 induction was also blunted in heterozygote organoids. (F) Design of two guide RNAs, e1 and e6, compatible with a repressive dCas9-KRAB. e1 is located at +363 bp, within the enhancer but outside the CnR-defined HNF4A binding site (Fig. 6D), whereas e6 is located at +781 bp within the HNF4A binding site (red box). (G) HNF4A+ organoids with an intact CDX2 enhancer (+/+) were transduced with lentivirus encoding dCas9-KRAS and e6, e1, or a nontargeting guide. Compared with the latter, the guide targeting the HNF4A binding site (e6) reduced Cdx2 mRNA expression, while the control e1 guide did not. (H) Model for increased HNF4A levels activating the 3′ shadow Cdx2 enhancer, hence inducing expression of CDX2, the TF that activates intestinal genes in stomach cells.