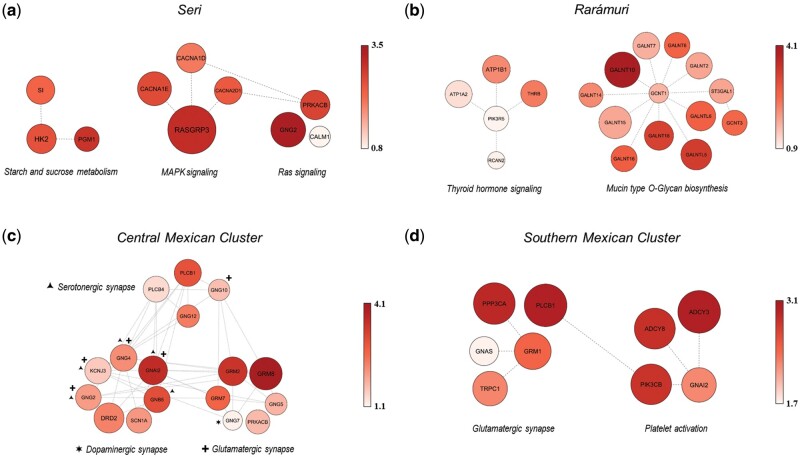
Fig. 3.
Representation of gene networks mediating polygenic adaptive events inferred for Seri, Rarámuri, and genetic clusters of Native Mexican populations identified according to fine-STRUCTURE analysis. (a) Gene networks belonging to the Starch and sucrose metabolism, Ras signaling, and MAPK signaling pathways showing significant footprints of positive selection in the SER population. (b) Gene networks belonging to the Thyroid hormone signaling and Mucin type O-Glycan biosynthesis pathways having experienced multiple selective events in the TAR population. (c) Highly interrelated gene networks belonging to the Glutamatergic, Serotonergic, and Dopaminergic synapse pathways showing widespread selection signatures in Central Mexican Cluster populations. Genes playing a role in other pathways other than the one in which they are represented are indicated with symbols: plus, Glutamatergic pathway; triangle, Serotonergic pathway; star, Dopaminergic synapse pathway. (d) Gene networks belonging to the Glutamatergic synapse and Platelet activation pathways having mediated the adaptive evolution of Southern Mexican Cluster populations. The color intensity of circles is displayed according to the reported color scale and, along with circles size, is proportional to the per-gene nSL representative scores used to perform gene-network analyses.