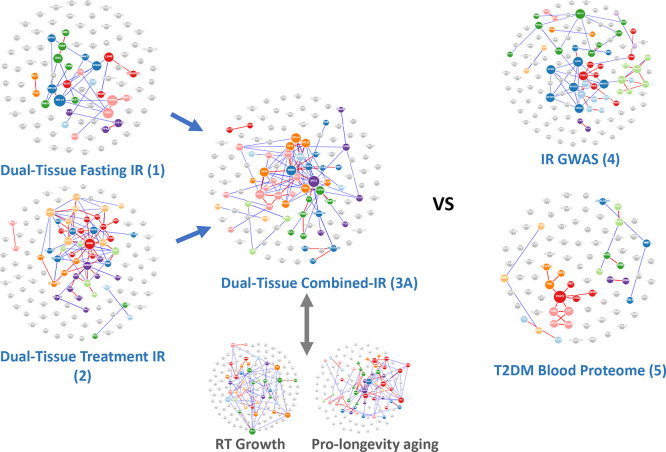
Appendix 1—figure 2. Network-based analysis of the gene lists from Table 1 analysed using protein-protein interaction data.
A notable characteristic of these gene lists was that prior knowledge using multi-omic interaction databases (http://apps.broadinstitute.org/genets) was unable to connect the members of the list, indicating that previously unknown information was contained within each list and our novel analysis. There were <3% overlap of genes across the signatures considered in this study (Table S1) and only two genes common between signatures 3A and 4 (INSR, GRB14), and these were not essential for the performance of the RNA-based insulin resistance-drug repurposing (IR-DR) signature. See Figure S3 and ‘Discussion’ for reference to additional human signatures beyond IR.