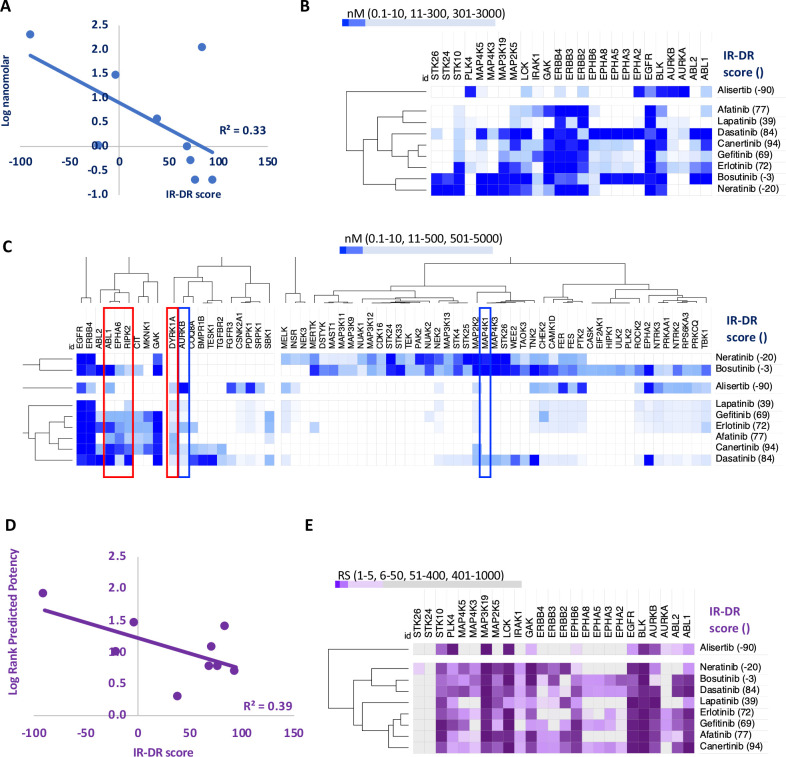
Figure 3. An analysis of the relationship between the insulin resistance-drug repurposing (IR-DR) score and laboratory-based pharmacological potency and selectivity or deep learning-based predictions of compound potency and selectivity.
(A) Inhibitory constants (nM) derived from laboratory assays against top-ranked targets for a series of epidermal growth factor receptor (EGFR) inhibitors. (B) Relationship between IR-DR score (100 = best score) and log potency against EGFR. (C) Expanded range of known targets, for at least one of the inhibitors, helps identify potentially positive (red box) and negative (blue box) off-target inhibitory actions. (D) Rank order score (RS, 1–19211) of predicted compound binding for all protein-coding genes using the DeepPurpose ML model; lab-validated targets feature in top 0.15% of target predictions. Log rank order (‘predicted potency’) for EGFR, over the protein-coding genome, partly predicts efficacy in IR-DR assay, confirming that the ML model matches the relationship observed using the laboratory pharmacology. (E) Using the predicted protein targets and the DeepPurpose rank order scores, it is possible to cluster positively acting ‘EGFR’ compounds from less active or negatively acting compounds.