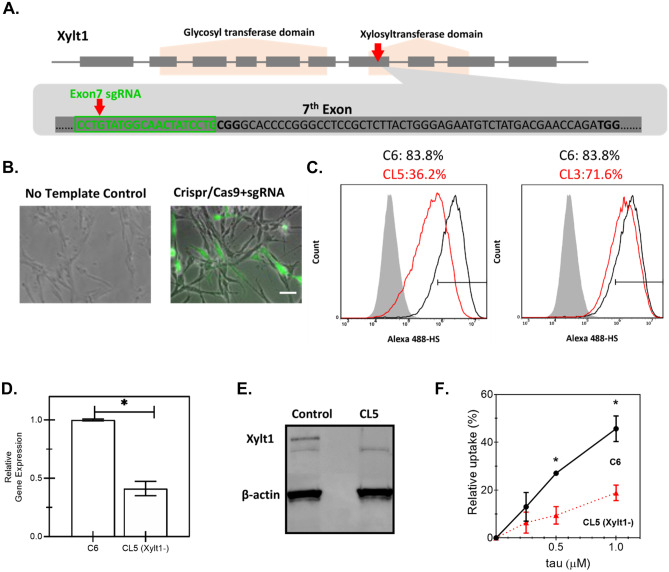
Fig. 4.
Construction of a Xylt1 knockdown C6 glioma cell line with CRISPR/Cas9 genome editing technology. A Schematic illustration of Xylt-1- gene of Rattus norvegicus, containing two functional domains and 10 exons. Single-guide RNA (sgRNA) was designed to target Exon7 of Xylt1 within the xylosyltransferase domain as indicated by the red arrow. The sgRNA sequence is highlighted in green. B Overlay of phase-contrast images with fluorescent images of transfected C6 cells with either the control or the pre-constructed CRISPR/Cas9 + sgRNA plasmid which uses GFP as a selection marker to enrich the cell population with higher levels of Cas9 and sgRNA expressions. Scale bar = 100 μm. C Quantification of heparan sulfate (HS) expression by flow cytometry of non-edited C6 (C6), clone 5 (CL5), or clone 3 (CL3), showing highest knockdown for CL5. Shaded histogram: negative control; black curve: parental C6 cells; red curve: either C6 single-cell clone (CL) 5 or 3, as indicated. Xylt1 gene knockdown in clone 5 (CL5) was verified by both quantitative reverse transcription-polymerase reaction (RT-qPCR) (D), where * indicates p < 0.05 between different test conditions, and western blot analysis using Xylt1 antibody (E). F The relative uptake of tau in C6 parental cells (filled circles) compared with CL5 cells (filled triangles) was quantified by flow cytometry after a 30-min incubation at the indicated concentrations. Error bars reflect the standard error, where * represents p < 0.05 for CL5 cells compared with parental (n = 2 independent biological replicates performed in duplicate)