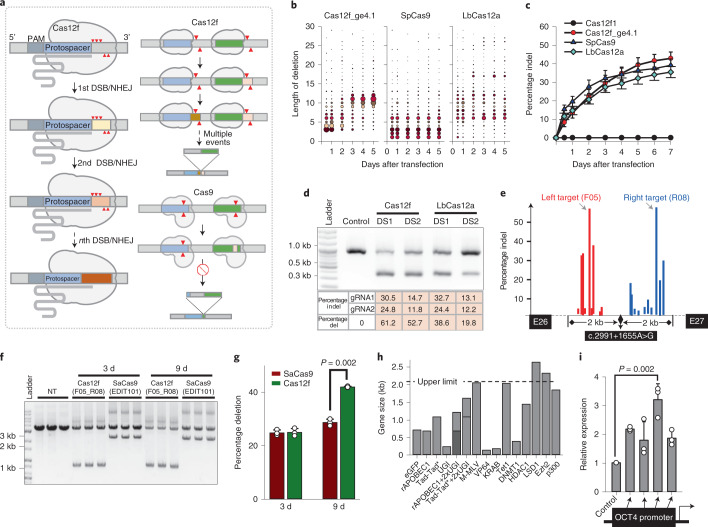
Fig. 3. Highly efficient correction of pathogenic mutations through AAV-delivered Cas12f.
a, A schematic illustration showing multiple chances for dsDNA cleavage and NHEJ cycles for the Cas12f system. Cleavage sites are marked with triangles and different colors indicate changes in the DNA sequences. The protospacer regions are colored sky-blue and green. b, Increased frequency of long deletion mutations over time in HEK293T cells transfected with CRISPR–Cas12f_ge4.1. c, Time-course of Cas12f-, SpCas9- and LbCas12a-induced indel frequencies in HEK293T cells transfected with plasmid vectors (n = 3). d. Comparison of Cas12f- and LbCas12a-mediated rates of exon 51 deletion from the human dystrophin gene in AC16 cells. The lower bands indicate the PCR amplicons of the exon 51-deleted locus. The intensity of the lower bands is indicative of the deletion efficiency. Deletion strategy 1 (DS1) and DS2 target identical loci for Cas12f and Cas12a. The data represent three experiments. e, Screening of targets for the deletion of the c.2991+1655A>G mutation from the CEP290 gene. f, Comparison of Cas12f- and SaCas9 (EDIT101)-mediated frequencies of deletion of the c.2991+1655A>G mutation. HEK293T cells (2 × 105) were seeded into 12-well plates, transduced with AAV2 harboring the Cas12f or SaCas9 system at 5.0 × 109 vector genomes (vg) ml−1, and harvested 3 and 9 d post-transduction. NT, nontransduction. The data represent three experiments. g, Quantitative analysis of deletion frequencies using RT-qPCR. Percentage deletion indicates the percentage ratio of PCR amplicons containing the deletion versus intact amplicons (n = 3). P values were derived using a two-sided Welch’s t-test. h, Possible applications of Cas12f using an AAV delivery system. For AAV delivery of vector constructs harboring the Cas12f sequence with two nuclear localization signals, a BGH poly(A) signal, and an XTEN linker sequence under the control of an EF-1α core promoter and a ge4.1 sequence under the control of a U6 promoter, a protein encoded by a gene ≤2.1 kb in size could be fused to Cas12f. i, Application of dCas12f-VP64 to CRISPRa. The fusion protein guided by gRNAs (ge4.1) targeting promoter regions of OCT4 led to transcriptional activations in HEK293T cells (n = 3). P values were derived using a two-sided Student’s t-test. All error bars represent the s.d.