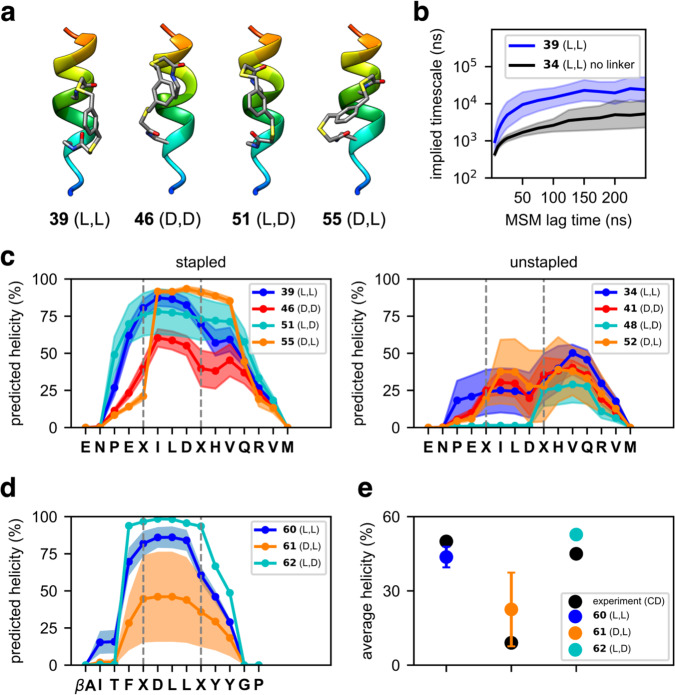
Fig. 6. Molecular dynamics simulation.
a Energy-minimized structures of 1,3-benzenedimethanethiol crosslinked Axin peptides 39, 46, 51 and 55. b The slowest implied timescale of peptides 39 and 34 (no linker) shown as a function of MSM lag time. c Predicted per-residue helicity profiles of stapled and unstapled Axin peptides. d Predicted per-residue helicity profiles of HIV C-CA binding peptides 60–62. e Comparison of experimental (CD) and predicted average helicities of peptides 60–62. Data in panels b–e are presented as mean values (dark lines) ± SEM (shaded regions) estimated from a bootstrap procedure where five MSMs (n = 5) were constructed by sampling the input trajectory data with replacement. .