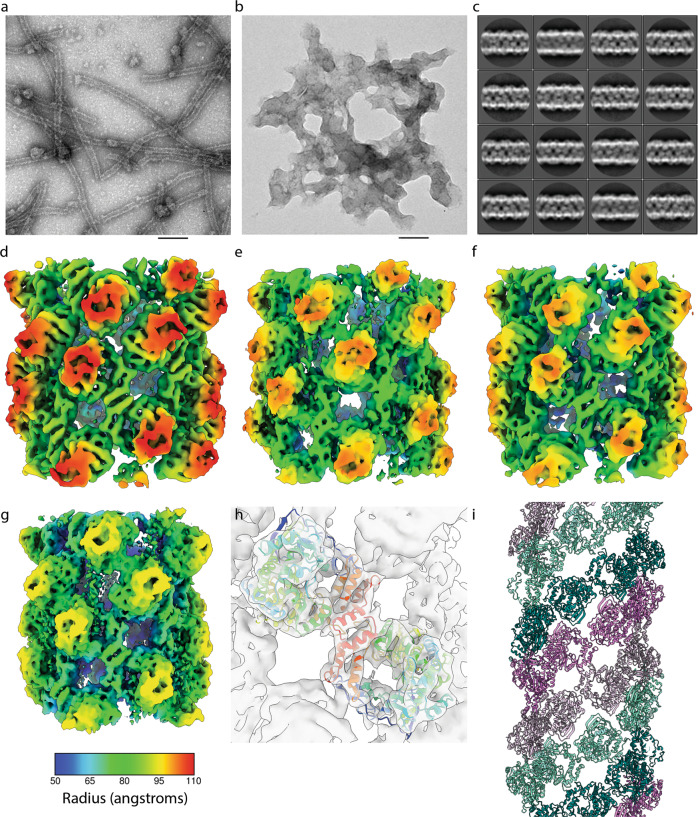
Fig. 4. Structure of 3Dpol fibrils.
Negative stain TEM of a sample from a polymerase activity assay in the presence of glutaraldehyde after 30-minute incubation. a 3Dpol-WT forms visible fibrils, b 3Dpol-GNN does not show any distinct fibril structure formation. Scale bar 100 nm. c 2D classification and averaging of fibril section images from cryo-EM data shows variation in fibril diameter. d–g Helical reconstruction led to the calculation of density maps for nine classes of fibril at 7.3–9.5 Å resolution, four representative maps are illustrated here coloured according to radius (see key below panel g). d The broadest fibril reconstruction; B1 had a diameter of 22.3 nm, C2 symmetry, a helical twist of 39o and an axial rise per subunit of 27 Å. e Conformation B2 has a more open structure, is also C2 symmetric and has a diameter of 21.7 nm, twist of 40o and rise of 31 Å. f N3 is a narrow (Ø = 21.5 nm) and tightly packed fibril which may also be described by a 1-start helical symmetry with a twist of -158.8o and axial rise of 15 Å. g N9 is a narrow, open fibril structure having a diameter of 20 nm, twist of -158.6° and axial rise of 17.4 Å. h Fitting of the X-ray structure for 3Dpol-WT in complex with template primer RNA and ATP (PDB: 2EC0) into each density map shows that these fibrils are two-start helical lattices comprising protofilaments of 3Dpol dimers. i Protofilament one is coloured violet/thistle, while protofilament two is coloured teal/aquamarine, two shades are used in each protofilament to highlight how 3Dpol monomers form ribbons of dimers.