Abstract
Accurate prostate cancer screening is imperative for reducing the risk of cancer death. Ultrasound imaging, although easy, tends to have low resolution and high inter-observer variability. Here, we show that our integrated machine learning approach enabled the detection of pathological high-grade cancer by the ultrasound procedure. Our study included 772 consecutive patients and 2899 prostate ultrasound images obtained at the Nippon Medical School Hospital. We applied machine learning analyses using ultrasound imaging data and clinical data to detect high-grade prostate cancer. The area under the curve (AUC) using clinical data was 0.691. On the other hand, the AUC when using clinical data and ultrasound imaging data was 0.835 (p = 0.007). Our data-driven ultrasound approach offers an efficient tool to triage patients with high-grade prostate cancers and expands the possibility of ultrasound imaging for the prostate cancer detection pathway.
Subject terms: Prostate, Computational science
Introduction
Prostate cancer is one of the most commonly diagnosed cancers in elderly men1. Ultrasound imaging is widely used in prostate cancer screening because it is nonionizing, low-cost, and safe. However, its low resolution and high inter-observer variability deteriorate the accuracy of ultrasound diagnosis2. Prostate cancer is a heterogeneous disease that ranges from indolent to aggressive3. Pathological grading is considered an important factor in predicting the prognosis of prostate cancer patients4. High-grade cancer tends to metastasize and is usually considered a castration-resistant prostate cancer5. On the other hand, older men with low-grade cancer who undergo treatment may experience complications without reducing their risk of dying from prostate cancer6. Accurate diagnosis through prostate cancer screening enables optimization of cancer management. Since pathological cancer grading determines therapeutic strategies, it is necessary not only to detect the cancer, but also to estimate the pathological grading, such as the Gleason score7, which would be desirable to know even during ultrasound examination.
Artificial intelligence (AI) technologies, including deep learning algorithms, are gaining extensive attention due to their excellent performance in image classification and object detection. Recently, these algorithms have been useful tools in the analysis of medical images of various cancers, such as breast cancers8, brain tumors9, lung cancers10, esophageal cancers11, skin malignancies12, and prostate cancers13–15. The robust performance of the deep learning algorithm indicates its potential clinical use for screening on computed tomography (CT) images of patients with suspected coronavirus disease 2019 (COVID-19) pneumonia16,17. Furthermore, deep learning techniques have also been applied to ultrasound imaging. Li et al. reported that their deep convolutional neural network model showed similar sensitivity in identifying thyroid cancer compared with skilled radiologists18. Gu et al. proposed an automated three-dimensional segmentation approach using deep learning on ultrasound images for breast cancer19. In the field of prostate ultrasound image analysis, deep learning techniques enable accurate automatic segmentation20. Furthermore, several studies proposed a deep learning framework to detect prostate cancer using contrast-enhanced ultrasound images21,22. However, it is highly challenging to estimate pathology-level cancer grading using deep learning based on ordinary ultrasound images. In this study, we aim to estimate pathological high-grade cancer using ordinary ultrasound images and limited clinical data.
Results
Image and patient characteristics
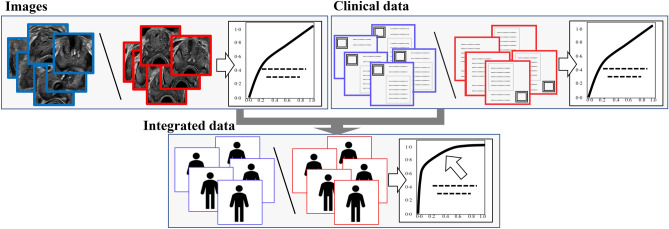
A flowchart of the study is presented in Fig. 1. We evaluated prediction accuracies for prostate cancer using the following data sets: still ultrasound image data (left upper box), clinical data (age and prostate-specific antigen [PSA]) (right upper box), and integrated data (ultrasound image data, total prostate volume [TPV] derived from ultrasound images, PSA density [PSAD], age, and PSA) (lower box).
Figure 1.
Flowchart of our study. Deep learning analysis for ultrasound images (upper left images), clinical data (upper right images), and integrated data (lower images).
Table 1 shows 691 cases (2676 images) that underwent ultrasound-guided prostate biopsy (systematic biopsy and magnetic resonance imaging [MRI]-targeted biopsy) at the Nippon Medical School Hospital. The median age (interquartile range) of all cases was 71 (65–76) years, and that of the cancer cases was 72 (66–77) years. Patients with cancer were significantly older than those without cancer (p < 0.001). The median PSA value of all the cases was 8.3 (5.8–14.0) ng/mL, and that of the cancer cases was 9.5 (6.7–20.5) ng/mL. The PSA level was significantly higher in cancer cases than in non-cancer cases (p = 0.002). The median TPV of all the cases was 35.0 (25.8–50.6) cm3, and that of the cancer cases was 30.9 (23.0–42.6) cm3. TPV was significantly lower in cancer cases than in non-cancer cases (p < 0.001). The median PSAD of all cases was 0.245 (0.138–0.482) ng/mL/cm3, and that of the cancer cases was 0.352 (0.207–0.681) ng/mL/cm3. PSAD was significantly higher in cancer cases than in non-cancer cases (p < 0.001). The number of biopsy Gleason scores was 6 (47 cases), 7 (215 cases), 8 (79 cases), 9 (94 cases), and 10 (1 case). High-grade cancer (Gleason score ≥ 8) accounted for 39.9% of the cancer cases.
Table 1.
Patient characteristics.
| Total | Cancer cases | Non-cancer cases | p value | |
|---|---|---|---|---|
| Cases, n (images, n) | 691 (2676) | 436 (1691) | 255 (985) | – |
| Age (years) | ||||
|
Median (IQR) Mean (SD) |
71, 65–76 69.9 ± 8.59 |
72, 66–77 71.3 ± 8.18 |
69, 62–74 67.4 ± 8.71 |
< 0.001 |
| PSA (ng/mL) | ||||
|
Median (IQR) Mean (SD) |
8.3, 5.8–14.0 128.9 ± 1034.7 |
9.5, 6.7–20.5 198.9 ± 1297.5 |
6.6, 4.9–10.4 9.20 ± 10.38 |
0.002 |
| TPV (cm3) | ||||
|
Median (IQR) Mean (SD) |
35.0, 25.8–50.6 42.8 ± 29.1 |
30.9, 23.0–42.6 37.1 ± 27.4 |
45.9, 32.5–62.0 52.5 ± 29.2 |
< 0.001 |
| PSAD (ng/mL/cm3) | ||||
|
Median (IQR) Mean (SD) |
0.245, 0.138–0.482 2.85 ± 18.1 |
0.352, 0.207–0.681 4.39 ± 22.6 |
0.140, 0.0939–0.214 0.201 ± 0.288 |
< 0.001 |
| Gleason score, n | ||||
| 6 | 47 | 47 | – | – |
| 7 | 215 | 215 | ||
| 8 | 79 | 79 | ||
| 9 | 94 | 94 | ||
| 10 | 1 | 1 | ||
PSA prostate-specific antigen, TPV total prostate volume, PSAD PSA density, IQR interquartile range, n number, SD standard deviation.
Classification of prostate cancers (n = 691)
Image-level classification using a deep neural network
We performed two sets of deep learning analyses using different labels. First, we applied a deep neural network, Xception23, to ultrasound image data with labels of the cancer group (positive, Gleason score ≥ 6) and the non-cancer group (negative). The area under the curve (AUC) of the cancer classification was 0.693 (95% confidence interval [CI] 0.640–0.746) (Table 2). Next, we applied the deep neural network to the same ultrasound image data with labels of the high-grade cancer group (positive, Gleason score ≥ 8) and the others (negative). The AUC of the high-grade cancer classification was 0.723 (95% CI 0.659–0.788) (Table 2). Note that we only used ultrasound images for these classifications. Supplementary Fig. S1 shows the receiver operating characteristic (ROC) curves for the classification accuracy.
Table 2.
AUCs of the cancer grading classification (n = 691).
| Images | Clinical data | Image integration | Data integration | p value* | |
|---|---|---|---|---|---|
| Cancer classification |
Deep learning 0.693 (95% CI 0.640–0.746) |
Ridge 0.671 (95% CI 0.563–0.779) |
Deep learning + ridge 0.774 (95% CI 0.680–0.868) |
Deep learning + ridge 0.789 (95% CI 0.697–0.880) |
0.104 |
|
Lasso 0.671 (95% CI 0.562–0.779) |
Deep learning + Lasso 0.774 (95% CI 0.680–0.868) |
Deep learning + Lasso 0.779 (95% CI 0.686–0.873) |
0.141 | ||
|
SVM 0.702 (95% CI 0.598–0.806) |
Deep learning + SVM 0.776 (95% CI 0.683–0.870) |
Deep learning + SVM 0.807 (95% CI 0.719–0.894) |
0.051 | ||
| High-grade cancer classification |
Deep learning 0.723 (95% CI 0.659–0.788) |
Ridge 0.675 (95% CI 0.564–0.786) |
Deep learning + Ridge 0.816 (95% CI 0.725–0.908) |
Deep learning + Ridge 0.822 (95% CI 0.736–0.908) |
0.012 |
|
Lasso 0.665 (95% CI 0.553–0.777) |
Deep learning + Lasso 0.816 (95% CI 0.724–0.907) |
Deep learning + Lasso 0.824 (95% CI 0.737–0.911) |
0.009 | ||
|
SVM 0.691 (95% CI 0.582–0.801) |
Deep learning + SVM 0.816 (95% CI 0.725–0.908) |
Deep learning + SVM 0.835 (95% CI 0.753–0.916) |
0.007 |
The bold text indicates the highest value of AUCs.
AUC area under the curve, CI confidence interval, SVM support vector machine.
*AUC of clinical data versus that of data integration.
Case-level classification based on clinical data
We also applied logistic regression and support vector machine (SVM)24,25 analyses to the clinical data with the aforementioned labels: (1) cancer classification label: cancer (Gleason score ≥ 6) or non-cancer group, (2) high-grade cancer classification label: high-grade cancer group (Gleason score ≥ 8) or the other. Age and PSA were used as clinical data in this study because these clinical data are known to be important factors in the screening of prostate cancers26. Table 2 presents the AUCs for each classification. The AUC of the cancer classification was 0.702 (95% CI 0.598–0.806), and that of the high-grade cancer classification was 0.691 (95% CI 0.582–0.801) (SVM). Note that we used only clinical data for this classification. Supplementary Fig. S2 shows the ROC curves for these classifications.
Case-level classification using an image integration approach
Next, we applied logistic regression and SVM to three ultrasound image data pre-analyzed by deep learning. We selected the ultrasound images with the top three highest probabilities (|Pdl − 0.5|, Pdl: predicted probability of the deep learning classification) in every case. Table 2 shows the AUCs. Supplementary Fig. S3 shows the ROC curves for these classifications. The highest value of AUCs in cancer classification was 0.776 (95% CI 0.683–0.870) (SVM), and that in the high-grade cancer classification was 0.816 (95% CI 0.725–0.908).
Case-level classification using a data integration approach
Finally, we applied logistic regression and SVM analyses to the integrated data (three ultrasound image data pre-analyzed by deep learning, TPV, PSAD, age, and PSA). We also selected the ultrasound images with the top three highest probabilities in every case. Table 2 shows the AUCs. Supplementary Fig. S4 shows the ROC curves for these classifications. In the integrated data, the highest value of AUCs in cancer classification was 0.807 (95% CI 0.719–0.894) (SVM), and that in the high-grade cancer classification was 0.835 (95% CI 0.753–0.916) (SVM). Figure 2 shows the ROC curves for the high-grade cancer classification of clinical data without data derived from ultrasound images and with that derived from ultrasound images (integrated data). The AUC of the integrated data was significantly higher than that of the clinical data (0.691 [95% CI 0.582–0.801]) in high-grade cancer classification (p = 0.007).
Figure 2.
ROC curves of high-grade cancer classification (n = 691: systematic biopsy and MRI-targeted biopsy cases). Blue line: ROC curve of the clinical data without ultrasound images. Red line: ROC curve of clinical data with ultrasound images (integrated data). Light blue area: 95% CI for the ROC curve of the clinical data without ultrasound images. Light red area: 95% CI for ROC curve of clinical data with ultrasound images (integrated data). ROC receiver operating characteristic, AUC area under the curve, CI confidence interval.
Classification of prostate cancers in cases of systematic biopsy (n = 532)
To eliminate selection bias based on MRI, we selected 532 cases of systematic biopsy without MRI-targeted biopsy. In image-level classification, the AUC of the cancer classification was 0.670 (95% CI 0.607–0.733), and that of the high-grade cancer classification was 0.732 (95% CI 0.658–0.807). We also applied logistic regression and SVM24,25 analyses based on the clinical data. The AUC of the cancer classification was 0.639 (95% CI 0.511–0.766) (SVM), and that of the high-grade cancer classification SVM was 0.665 (95% CI 0.535–0.796). Next, we applied logistic regression and SVM anlyses to three ultrasound image data pre-analyzed by deep learning. The AUC of the cancer classification was 0.722 (95% CI 0.602–0.841) (SVM), and that of the high-grade cancer classification was 0.814 (95% CI 0.708–0.920) (SVM). Finally, logistic regression and SVM anlyses were applied to the integrated data. The AUC of the cancer classification was 0.750 (95% CI 0.636–0.863) (SVM), and that of the high-grade cancer classification was 0.831 (95% CI 0.738–0.924) (SVM) (Supplementary Table S1). Supplementary Figs. S5, S6, S7, and S8 show the ROC curves for the classification accuracies. Figure 3 shows the ROC curves for the high-grade cancer classification of clinical data without data derived from ultrasound images and those derived from ultrasound images (p = 0.013). In AUCs, a tendency similar to that in “Classification of prostate cancers (n = 691)” was observed.
Figure 3.
ROC curves of high-grade cancer classification (n = 532: only systematic biopsy cases). Blue line: ROC curve of the clinical data without ultrasound images. Red line: ROC curve of clinical data with ultrasound images (integrated data). Light blue area: 95% CI for the ROC curve of the clinical data without ultrasound images. Light red area: 95% CI for ROC curve of clinical data with ultrasound images (integrated data). ROC receiver operating characteristic, AUC area under the curve, CI confidence interval.
Prostate ultrasound images of top five cases corresponding to histological cancer grading
Figure 4 shows the prostate ultrasound images of the top five cases according to pathological cancer grading based on the predicted probabilities of each class. The upper images show ultrasound images of high-grade cancer cases using our proposed method. The middle section shows images of the low-grade cancer cases, and the lower images show non-cancer cases. Expert urologists analyzed these images as follows. Ultrasound images of high-grade prostate cancer (upper images) show an asymmetric prostatic lobe, an unclear boundary of the prostatic capsule, and extensive hypoechoic areas. In contrast, ultrasound images of low-grade cancer (middle images) show an intensive hypoechoic area, while maintaining the clear boundary of the prostatic capsule and symmetric prostatic lobe. On the other hand, ultrasound images of non-cancer cases (lower images) show a symmetric prostatic lobe and an isoechoic area with a clear boundary of the prostatic capsule.
Figure 4.
Prostate ultrasound images of top 5 cases corresponding to pathological cancer grading. Upper images: high-grade cancer cases. Middle images: low-grade cancer cases. Lower images: non-cancer cases. PP: normalized predicted probability.
A representative case with saliency map using explainable deep learning model
Supplemental Fig. S9 shows a representative case with saliency map using explainable deep learning model. We applied gradient-weighted class activation mapping (Grad-CAM) to construct a saliency map27,28.
Discussion
We aimed to idenify pathological high-grade prostate cancer using ultrasound images and limited clinical data. We evaluated the prediction accuracies of the following different datasets: ultrasound image data, clinical data, and integrated data.
Several imaging examinations are performed complementary, each with its own strengths. For example, MRI contributes to the detection of significant prostate cancers. In recent years, several studies have reported the benefits of MRI prior to systematic biopsy29. Pellicer-Valero et al. proposed a fully automatic system based on deep learning with prostate MRI that can show cancer segmentation and cancer grading30. In their results, AUC of intermediate and high grade cancer detection (Gleason grade group ≥ 3) was 0.767 (ProstateX test data31) and 0.840 (Valencia Oncology Institute Foundation data30). On the othre hand, AUC when using our method of high grade cancer detection (Gleason grade group ≥ 4) was 0.835. The ROC curve showed sensitivity of 0.909 and specificity of 0.609 at a cut off value optimized for triage. Ultrasound imaging is widely used in prostate cancer screening globally because ultrasound can be easily performed at bedside. In this study, we showed that our data-driven ultrasound approach offers an efficient tool to triage patients with high-grade prostate cancers.
The main limitation of this study was that it was conducted at a single facility. It is known that high inter-observer variability deteriorates the accuracy of ultrasound diagnosis. However, we analyzed over 2500 ultrasound images. In addition, we applied an augmentation technique and transferred learning based on ImageNet32. In the future, we will obtain a validation set for other facilities. Furthermore, a full three-dimensional ultrasound image analysis might be able to improve the prediction accuracy in order not to miss small cancer lesions. Although further investigation should be conducted in order to reinforce our results, we hope that our method will contribute to the accurate diagnosis of prostate cancer.
Deep learning algorithms have achieved great success in medical image analyses owing to the high affinity between neural networks and images. Integrated analysis of medical multimodal data is a key factor driving practical technology in the next stage. Even if the predictive power of each datum is insufficient, data integration can improve the predictive power by appropriate machine learning techniques. Identifying the appropriate combination of data is important for better use of the data stored in the hospital. In this study, the image selection itself was performed quantitatively. This is an important step towards more accurate and integrated medical AI. Our study expands the possibility of ultrasound imaging. Recognizing pathological high-grade cancers during ultrasound procedures improves cancer management, significantly reduces mental burden on the patient, and leads to improved quality of life.
Materials and methods
Study population
Our study included 772 consecutive patients and 2899 ultrasound images acquired between November 2017 and June 2020. The patients underwent ultrasound-guided prostate biopsy at Nippon Medical School Hospital in Tokyo, Japan. The study profile is shown in Fig. 5. We excluded cases with a transperineal biopsy of the prostate (eight cases), a history of post intravesical bacillus Calmette-Guerin therapy (two cases), and others (insufficient saved image and data: 71 cases). We evaluated 2676 ultrasound images (691 cases) via a transrectal approach using deep learning analysis. A systematic biopsy was performed in 532 patients. A combination of both MRI-targeted and systematic biopsies was performed in 159 cases. We divided these images into training data (November 2017 to December 2019: 590 cases, 2,299 images) and test data (January 2020–June 2020: 101 cases, 377 images). We determined the hyperparameters using only the training data (Supplementary Table S2). This study was confirmed and approved by the Institutional Review Boards of the Nippon Medical School Hospital (reference 28-11-663) and RIKEN (reference Wako3 29-14). Informed consent was obtained from all patients. All methods were carried out in accordance with relevant guidelines and regulations.
Figure 5.
Study profile.
Ultrasound and biopsy procedure
Ultrasound images of prostate glands were saved at four locations (base, mid, apex-mid, and apex). The prostate volume was calculated for each case. Subsequently, a prostate biopsy was performed. Figure 6 shows systematic prostate biopsy sites at each location (number of biopsy sites): lateral apex (2), parasagittal apex (2), lateral apex-mid (2), parasagittal apex-mid (4), lateral mid (2), and lateral base (2). In 532 cases, a systematic biopsy was performed. Furthermore, in 159 cases, a combination of both MRI-targeted and systematic biopsies was performed. The highest biopsy Gleason score on each ultrasound image was used as the label of the corresponding images. We gave different Gleason scores for each ultrasound image. We used an ultrasound system (Aplio i800; Canon Medical Systems, Tokyo, Japan) with a 6 MHz transrectal probe (PVT-770 RT; Canon Medical System, Tokyo, Japan). All still ultrasound images were saved in Digital Imaging and Communications in Medicine (DICOM) format.
Figure 6.
Prostate biopsy sites (systematic biopsy). Red needles indicate the prostate biopsy sites in our study.
Pathological cancer grading
Two pathologists reviewed each biopsy core and reported cancer with an assigned Gleason score. Prostate cancer was diagnosed pathologically based on the International Society of Urological Pathology grading33. Pathologists diagnosed all cases independently and then reached a consensus.
Ultrasound images
All DICOM ultrasound images were converted into JPEG images. We then extracted a rectangular region of the prostate from the images. This rectangular region included proximate tissues, such as the prostatic capsular vessels, pelvic fascia, and rectum. We then adjusted these images to a size of 256 × 256 pixels. We applied an augmentation technique, including a parameter of zoom range for deep learning analysis. We provided positive or negative labels to these datasets based on the pathological grading. We defined (1) labels for the cancer group (Gleason score ≥ 6) or the other. We also defined (2) labels for the high-grade cancer group (Gleason score ≥ 8) or the other.
Classification using a deep neural network
First, we tested three deep convolutional neural network models, Xception23, inceptionV334, and VGG1635 which were pre-trained on ImageNet with classification layers adapted to our labels32. We selected Xception in this study because it exhibited the most precise performance for ultrasound image classification. We divided all images into test and training data based on the date of ultrasound evaluation for each case (Fig. 5). To evaluate the classification by the deep neural network, we constructed an ROC curve with the corresponding AUC. This study employed the RIKEN AIP Deep Learning Environment (RAIDEN) supercomputer system for all computations.
Machine learning analysis for the case-level prediction
First, we applied logistic regression (ridge and lasso) and SVM anlayses only to the clinical data (age and PSA) to classify the case data. Subsequently, we applied these algorithms to three ultrasound image data pre-analyzed by deep learning (predicted probability of deep learning classification). Comparison among the different number of image sets showed that the AUC was the highest with the top three highest probabilities (|Pdl − 0.5|, Pdl: predicted probability of deep learning classification) (Supplementary Table S3). We, therefore, selected ultrasound images with those three ultrasound images in every case. We used only cases with more than three images in this case-level prediction. 678 cases were remained for case-level classification. We also divided these case data into training and test sets, as described in “Study population”. Finally, we applied these algorithms to the integrated data: three ultrasound image data pre-analyzed by deep learning, TPV, PSAD, age, and PSA. We constructed an ROC curve using the corresponding AUC. We used R software for the analysis, using the glmnet package (version 2.0.16) for the ridge and lasso regression and the e1071 package (version 1.7.0) for the SVM. Calculations were performed automatically using the software packages.
Prostate ultrasound images corresponding to histological cancer grading
We evaluated prostate ultrasound images corresponding to pathological cancer grading based on two types of labels: (1) the cancer classification label and (2) the high-grade cancer classification label. We selected the top five highest predicted probability cases from the high-grade cancer logistic regression (ridge) as representative cases corresponding to the high-grade cancer group. We selected the top five lowest predicted probability cases from the cancer logistic regression as representative cases corresponding to the non-cancer group. We defined the top five highest predicted probability cases of cancer without high-grade cancer as the low-grade cancer group.
Saliency map
We applied Grad-CAM to construct a saliency map for deep learning analysis27,28. Grad-CAM is a technique used to produce visual explanations of decisions made by convolutional neural networks.
Statistical analysis
We compared the characteristics of patients who were cancer cases or non-cancer cases using the Wilcoxon rank-sum test for continuous data. The ROC curves were constructed and compared using the ‘pROC’ (version 1.13.0) package in R36. All reported p values were two-sided, with the level of statistical significance set at p < 0.05.
Supplementary Information
Author contributions
Conceptualization: J.A. and Y.Y.; methodology: Y.Y.; software: Y.N. and H.M.; formal analysis: Y.N.; investigation: J.A., Y.N., H.M., T.S., S.K., H.M., M.Y., Y.E., H.T., Y.T., R.Y., and Y.Y.; resources: Y.K. and Y.Y.; data curation: J.A. and Y.N.; writing: J.A., Y.N., and Y.Y.; supervision: G.K., Y.K., and Y.Y.; project administration: Y.Y.; funding acquisition: J.A. and Y.Y. All authors reviewed the manuscript.
Funding
This study was supported by the 14th JUA Grant for Young Researchers, a research Grant from the Nippon Medical School Alumni Association, the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (MEXT KAKENHI, Grant numbers: 18H05301 and 21K18102), and the Moonshot Research and Development Program ‘Ultra-early disease prediction and intervention’, Japan Science and Technology Agency (JPMJMS2022-9).
Data availability
The clinical datasets used were collected at the Nippon Medical School Hospital. They are not publicly available, and restrictions apply to their use.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jun Akatsuka and Yasushi Numata.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-04951-3.
References
- 1.Sung H, et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Mitterberger M, et al. Ultrasound of the prostate. Cancer Imaging. 2010;10:40–48. doi: 10.1102/1470-7330.2010.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltran H, Demichelis F. Prostate cancer: Intrapatient heterogeneity in prostate cancer. Nat. Rev. Urol. 2015;12:430–431. doi: 10.1038/nrurol.2015.182. [DOI] [PubMed] [Google Scholar]
- 4.Mottet N, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur. Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Yamoah, K. et al. Novel transcriptomic interactions between immune content and genomic classifier predict lethal outcomes in high-grade prostate cancer. Eur. Urol.S0302-2838(20), 30939–8 (2020). [DOI] [PMC free article] [PubMed]
- 6.Welch HG, Albertsen PC. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986–2005. J. Natl. Cancer Inst. 2009;101:1325–1329. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein JI. An update of the Gleason grading system. J. Urol. 2010;183:433–440. doi: 10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 8.Ehteshami Bejnordi B, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadeem, M. W. et al. Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges. Brain Sci.10, 118 (2020). [DOI] [PMC free article] [PubMed]
- 10.Ardila D, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 11.Tomita N, et al. Attention-based deep neural networks for detection of cancerous and precancerous esophagus tissue on histopathological slides. JAMA Netw. Open. 2019;2:e1914645. doi: 10.1001/jamanetworkopen.2019.14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto Y, et al. Automated acquisition of explainable knowledge from unannotated histopathology images. Nat. Commun. 2019;10:5642. doi: 10.1038/s41467-019-13647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulten W, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncol. 2020;21:233–241. doi: 10.1016/S1470-2045(19)30739-9. [DOI] [PubMed] [Google Scholar]
- 15.Ström P, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020;21:222–232. doi: 10.1016/S1470-2045(19)30738-7. [DOI] [PubMed] [Google Scholar]
- 16.Harmon SA, et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11:4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, et al. Deep learning-based triage and analysis of lesion burden for COVID-19: A retrospective study with external validation. Lancet Digit. Health. 2020;2:e506–e515. doi: 10.1016/S2589-7500(20)30199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: A retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20:193–201. doi: 10.1016/S1470-2045(18)30762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu P, et al. Automated 3D ultrasound image segmentation to aid breast cancer image interpretation. Ultrasonics. 2016;65:51–58. doi: 10.1016/j.ultras.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlando N, Gillies DJ. Automatic prostate segmentation using deep learning on clinically diverse 3D transrectal ultrasound images. Med. Phys. 2020;47:2413–2426. doi: 10.1002/mp.14134. [DOI] [PubMed] [Google Scholar]
- 21.Feng Y, et al. A deep learning approach for targeted contrast-enhanced ultrasound based prostate cancer detection. IEEE ACM Trans. Comput. Biol. Bioinform. 2019;16:1794–1801. doi: 10.1109/TCBB.2018.2835444. [DOI] [PubMed] [Google Scholar]
- 22.Sedghi A, et al. Improving detection of prostate cancer foci via information fusion of MRI and temporal enhanced ultrasound. Int. J. Comput. Assist. Radiol. Surg. 2020;15:1215–1223. doi: 10.1007/s11548-020-02172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chollet, F. Xception: Deep learning with depthwise separable convolutions. https://arxiv.org/abs/1610.02357 (Accessed 19 November 2021).
- 24.Vapnik V, editor. Statistical Learning Theory Ch. 12. Wiley; 1998. [Google Scholar]
- 25.Yamamoto Y, et al. Quantitative diagnosis of breast tumors by morphometric classification of microenvironmental myoepithelial cells using a machine learning approach. Sci. Rep. 2017;7:46732. doi: 10.1038/srep46732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam TBL, et al. EAU-EANM-ESTRO-ESUR-SIOG prostate cancer guideline panel consensus statements for deferred treatment with curative intent for localised prostate cancer from an international collaborative study (DETECTIVE study) Eur. Urol. 2019;76:790–813. doi: 10.1016/j.eururo.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Selvaraju, R.R. et al. Grad-CAM: Visual explanations from deep networks via gradient-based localization. IEEE Int. Conf. Comput. Vis. 618–626 (2017).
- 28.Akatsuka J, et al. Illuminating clues of cancer buried in prostate MR image: Deep learning and expert approaches. Biomolecules. 2019;9:673. doi: 10.3390/biom9110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callender T, Emberton M, Morris S, Pharoah PDP, Pashayan N. Benefit, harm, and cost-effectiveness associated with magnetic resonance imaging before biopsy in age-based and risk-stratified screening for prostate cancer. JAMA Netw. Open. 2021;4:e2037657. doi: 10.1001/jamanetworkopen.2020.37657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellicer-Valero, O. J.et al. Deep learning for fully automatic detection, segmentation, and Gleason Grade estimation of prostate cancer in multiparametric Magnetic Resonance Images. https://arxiv.org/abs/2103.12650 (Accessed 19 November 2021). [DOI] [PMC free article] [PubMed]
- 31.Armato SG, 3rd, et al. PROSTATEx challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images. J. Med. Imaging. 2018;5:044501. doi: 10.1117/1.JMI.5.4.044501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng, J. D. et al. ImageNet: A large-scale hierarchical image database. in 2009 IEEE. Conference on Computer Vision and Pattern Recognition, 248–255 (2009).
- 33.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am. J. Surg. Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 34.Szegedy, C. V. V., V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. https://arxiv.org/abs/1512.00567 (Accessed 19 November 2021).
- 35.Simonyan, K. Z. A. Very deep convolutional networks for large-scale image recognition. https://arxiv.org/abs/1409.1556 (Accessed 19 November 2021).
- 36.Robin X, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The clinical datasets used were collected at the Nippon Medical School Hospital. They are not publicly available, and restrictions apply to their use.