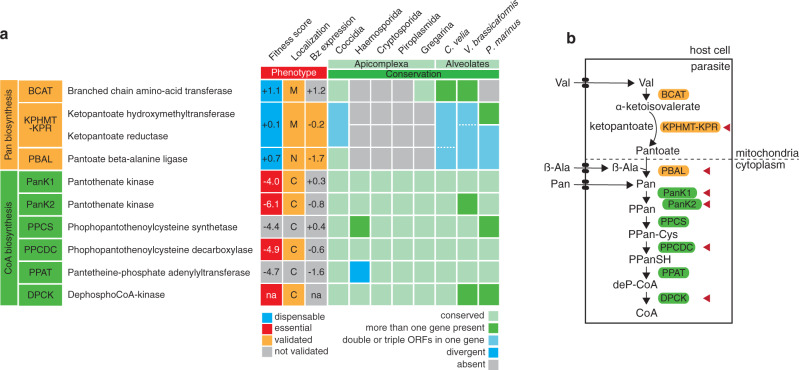
Fig. 1. Pan and CoA biosynthesis pathways in T. gondii.
a Table representing the fitness cost, localization, and stage expression of enzymes for pantothenate and coenzyme A biosynthesis in T. gondii, and conservation of the genes in other alveolates. Fitness scores from a genome-wide fitness screen are given (positive values indicate dispensability, while negative values suggest essentiality)15, together with dispensability/essentiality determined from this and previous studies (blue - dispensable, red - essential). Localizations, experimentally determined in this or previous studies, are given for all enzymes involved in pantothenate (Pan) or coenzyme A (CoA) biosynthesis (M mitochondrion, N nucleus, C cytosol), in gray when predicted but not experimentally validated. Log2 fold change in RNA levels from an RNA-seq analysis75 are provided (bradyzoite (Bz) expression levels relative to tachyzoites) and complemented by protein expression levels determined in this study (orange – validated experimentally). Conservation is determined by a BLAST search (e-value < E−04). Dotted lines represent the situation where single/fused genes are present, as well as one open reading frame (ORF) spanning all 3 genes. b Cartoon scheme of the Pan and CoA biosynthesis pathways. Valine (Val), β-Alanine (β-Ala) and Pan are likely salvaged through unknown transporters. Enzymes are in boxes. Red arrowheads highlight the enzymes under scrutiny in this manuscript. Abbreviations: PPan, 4′-phosphopantothenate; PPan-Cys, 4′-phospho-N-pantothenoyl cysteine; PPanSH, 4′-phosphopantetheine; deP-CoA, dephospho-CoA; CoA, coenzyme A.