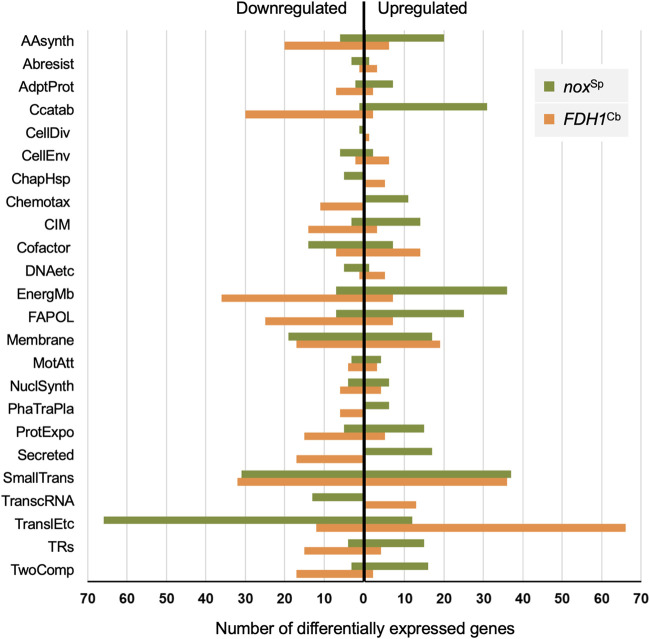
FIG 7.
Overexpression of noxSp and FDH1Cb generate antagonistic transcriptional profiles in P. aeruginosa. The 794 common genes that were found differentially regulated in both noxSp- and FDH1Cb-producing strains are classified according to the PseudoCAP functional categories (62) and grouped according to the direction of the regulation respective to their empty vector controls (up- or downregulated). The PseudoCAP categories are abbreviated as follows: AAsynth, amino acid biosynthesis and metabolism; ABresist, antibiotic resistance and susceptibility; AdptProt, adaptation and protection; CCatab, carbon compound catabolism; CellDiv, cell division; CellEnv, Cell wall/lipopolysaccharide (LPS)/capsule; ChapHsp, chaperones and heat shock proteins; Chemotax, chemotaxis; CIM, central intermediary metabolism; Cofactor, biosynthesis of cofactors, prosthetic groups, and carriers; DNAetc, DNA replication, recombination, modification and repair; EnergMb, energy metabolism; FAPOL, fatty acid and phospholipid metabolism; Membrane, membrane proteins; MotAtt, motility and attachment; NuclSynth, nucleotide biosynthesis and metabolism; PhaTraPla, related to phage, transposon, or plasmid; ProtExpo, protein secretion/export apparatus; Secreted, secreted factors (toxins, enzymes, alginate); SmallTrans, transport of small molecules; TranscRNA, transcription, RNA processing, and degradation; TranslEtc, translation, posttranslational modification, degradation; TRs, transcriptional regulators; TwoComp, two-component regulatory systems. Hypothetical and putative genes are excluded from the figure.