FIG 3.
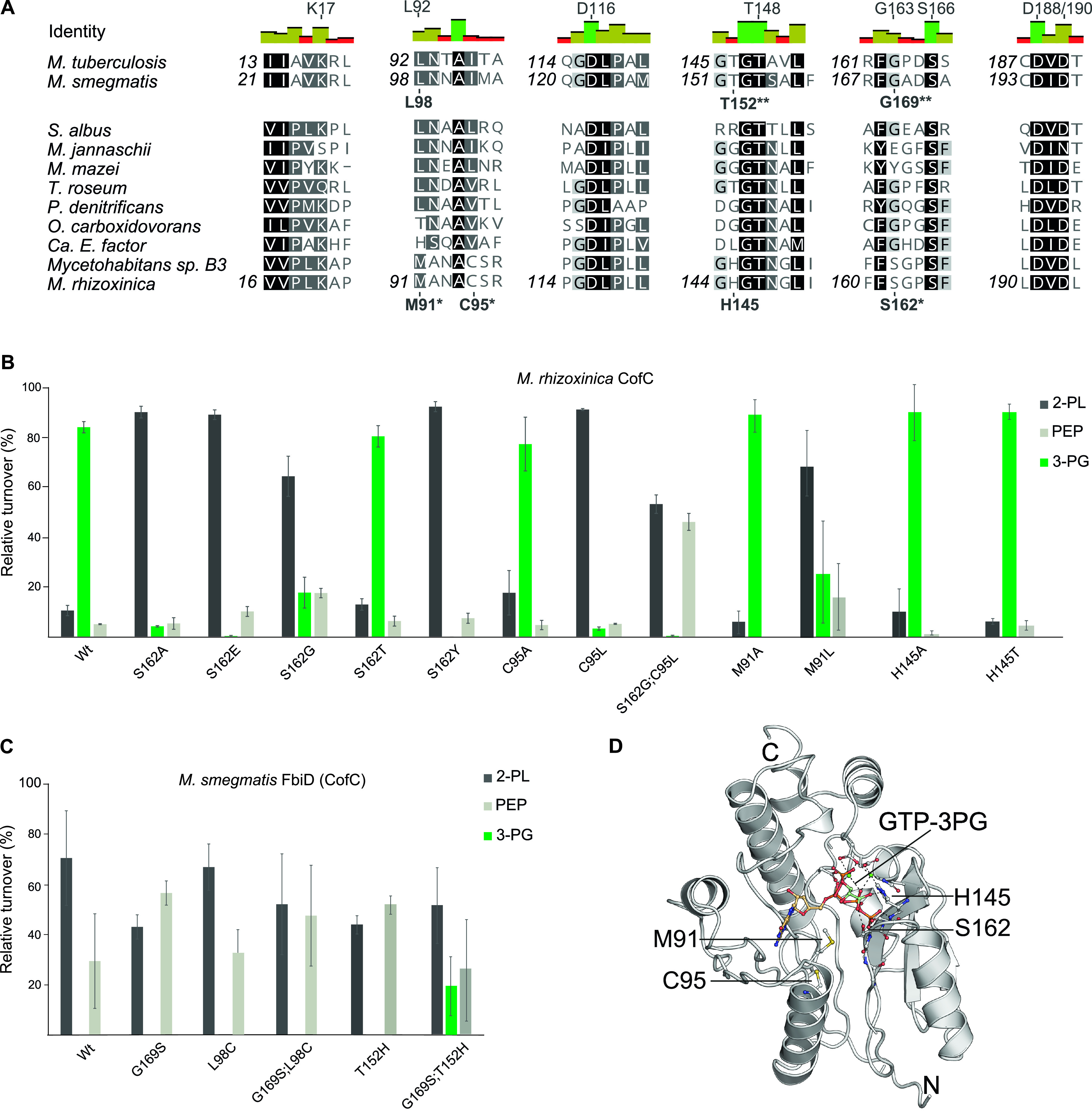
Residues determining substrate specificities of CofC homologs. (A) Multiple sequence alignment of CofC proteins from selected source organisms. Amino acids of Mtb-FbiD suggested previously to be involved in PEP binding residues (32) are indicated above the identity graph. Residues tested by mutagenesis are shown below sequences. Asterisks: crucial for 3-PG activation in Mrhiz-CofC, double asterisk: enabled 3-PG activation by Msmeg-FbiD. (B) Substrate specificity of Mrhiz-CofC after site-directed mutagenesis. Substitution of S162, C95, and M91 by residues occurring in 2-PL/PEP activating enzymes led to reduction or abolishment of 3-PG activation. (C) Substrate specificity of Msmeg-FbiD after site-directed mutagenesis. Gly169, Leu98, and Thr152 were exchanged by amino acids found in 3-PG activating homologs. While single mutations did not result in 3-PG activation, the combination G169S;T152H enabled 3-PG turnover. Error bars represent the standard deviation of three biological replicates. (D) Homology model of Mrhiz-CofC in complex with GTP (placed by molecular docking) and 3-PG (placed manually). For details, see Fig. S1 in the supplemental material.
