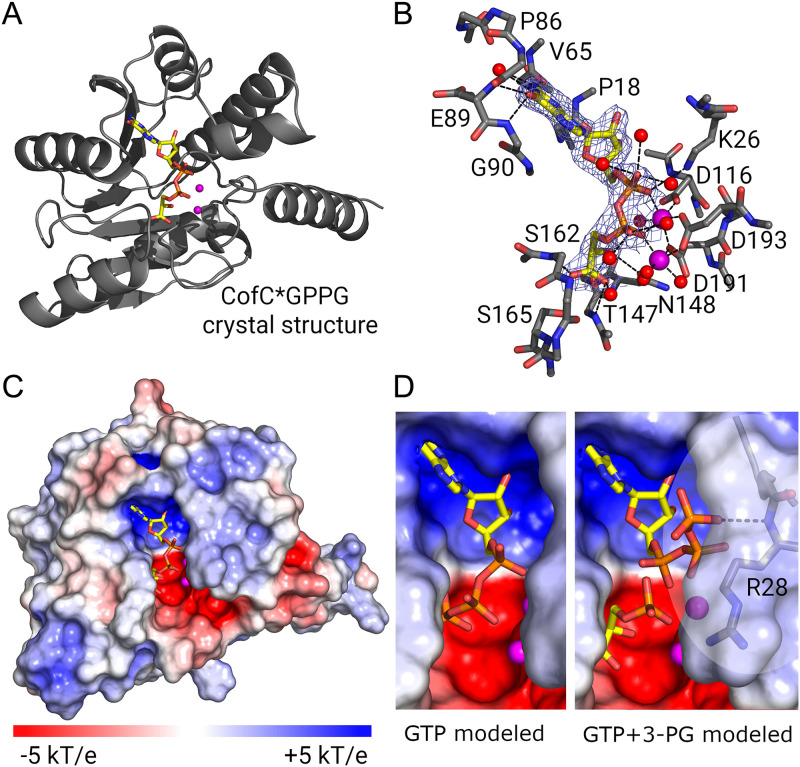
FIG 4.
Structures of MycB3-CofC in complex with GPPG (crystal structure) and educts (model). (A) Ribbon diagram of the crystal structure of CofC with its product GPPG. (B) Final electron density of GPPG (2FoFc at 1.5 σ) and H-bonds for GPPG binding. Side chains are only shown for residues contacting the ligand. (C) Electrostatic surface representation of the protein shows a negatively charged deep pocket for GPPG. Two Mg2+-ions allow binding of the charged phosphates of GPPG. The electrostatic surface potential was calculated using APBS. Red: negative charge; blue: positive charge. (D) Possible educt conformations. GTP could position its α- and β-phosphates where GPPG binds to the Mg2+-ions (left side). With 3-PG binding in the same mode as the 3-PG moiety of GPPG the β- and γ-phosphates of GTP have to rotate out of the binding pocket, though (right side). The phosphate moiety of 3-PG is well poised for nucleophilic attack on the α-phosphate of GTP via a trigonal bipyramidal transition state. Figures were produced with PyMOL. A window around R28 is transparent to show the potential H-bond of GTP to the amide nitrogen. Electrostatic surface representations were made using the APBS electrostatics plugin in PyMOL. Carbon: yellow, oxygen: red, nitrogen: blue, magnesium: magenta.