FIG 2.
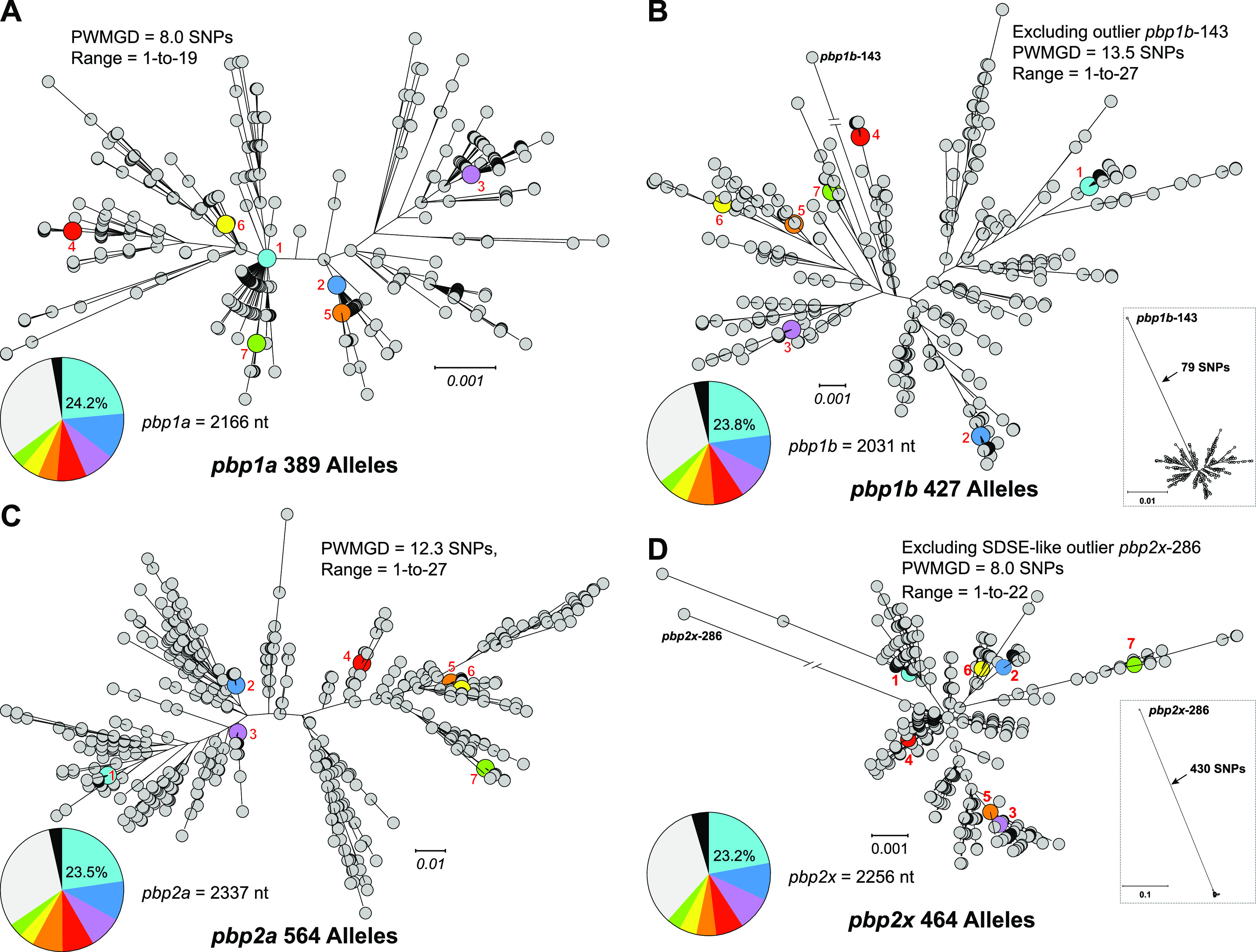
Genetic relationships among HMM PBP alleles. Relationships among alleles of each of the four HMM PBP genes were inferred by maximum likelihood and are illustrated as radial phylograms: pbp1a (A), pbp1b (B), pbp2a (C), and pbp2x (D). The pairwise mean genetic distance (PWMGD) in terms SNP differences between alleles is provided for each allele set. The inset trees (lower right) in panels B and D show the greater genetic distance of the recombinant outlier alleles pbp1b-143 and pbp2x-286 relative to the more closely related alleles of PBP1B and PBP2X, respectively. The seven most prevalent alleles for each of the PBPs are numbered and indicated in color. The inset pie charts (lower left) illustrate the relative abundance of the seven most prevalent alleles (gray = other alleles, black = undetermined). Among the isolates for which an allele was determined, the seven most prevalent alleles account for 66.7% (17,155/25,721) of pbp1a sequences, 66.9% (17,002/25,410) of pbp1b sequences, 67.8% (17,323/25,547) of pbp2a sequences, and 64.4% (16,281/25,265) of pbp2x sequences. In contrast, 30.6% (119/389) of pbp1a alleles were present in only a single sample in the strain cohort versus 30.4% (130/427) for pbp1b, 41.3% (233/564) for pbp2a, and 37.8% (175/464) for pbp2x.
