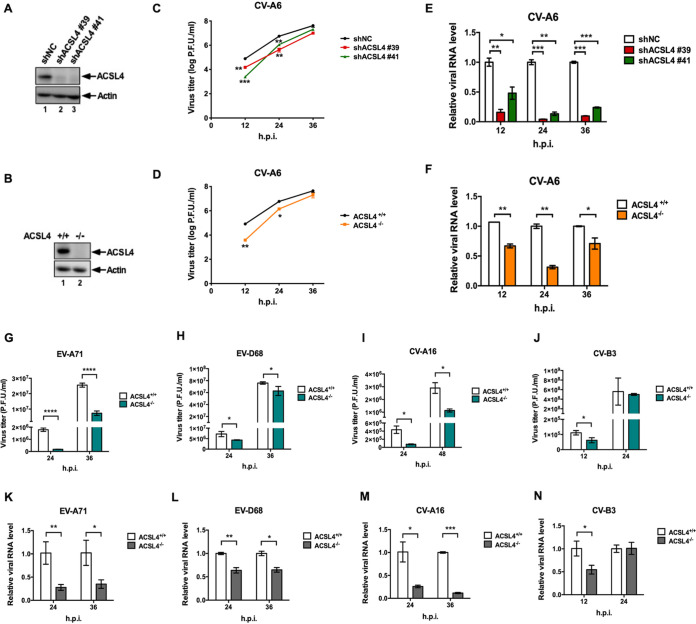
FIG 2.
ACSL4 is a broad host factor for enterovirus replication. RD cells were used to generate ACSL4 KD and ACSL4−/− cells. Western blots represent the expression of ACSL4 in ACSL4 KD cells (A) and in ACSL4−/− cells (B). (C) Viral yields of CV-A6 in ACSL4 KD cells. ACSL4 KD cells (shACSL4 #39 and shACSL4 #41) and negative-control cells (shNC) were infected with CV-A6 at an MOI of 0.01. Viruses were harvested at the indicated time points, and viral titers were determined by a plaque assay. (D, G, H, I, and J) ACSL4−/− and ACSL4+/+ cells developed using CRISPR-Cas9 editing were infected with CV-A6 at an MOI of 0.01 (D), EV-A71 at an MOI of 0.001 (G), EV-D68 at an MOI of 0.001 (H), CV-A16 at an MOI of 0.1 (I), or CV-B3 at an MOI of 0.001 (J). Viruses were harvested at the indicated time points, and viral titers were measured by a plaque assay. (E, F, K, L, M, and N) RNA quantification of enteroviruses in ACSL4 KD and ACSL4−/− cells. (E) ACSL4 KD and shNC cells were infected with CV-A6 at an MOI of 0.01. ACSL4−/− and ACSL4+/+ cells were infected with CV-A6 at an MOI of 0.01 (F), EV-A71 at an MOI of 0.001 (K), EV-D68 at an MOI of 0.001 (L), CV-A16 at an MOI of 0.1 (M), or CV-B3 at an MOI of 0.001 (N). Cells were harvested at the indicated time points. RNA extraction followed by quantitative reverse transcription PCR (qRT-PCR) was performed. Data are presented as means ± standard deviations (SD) from three independent experiments and analyzed using Student’s two-tailed unpaired t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.