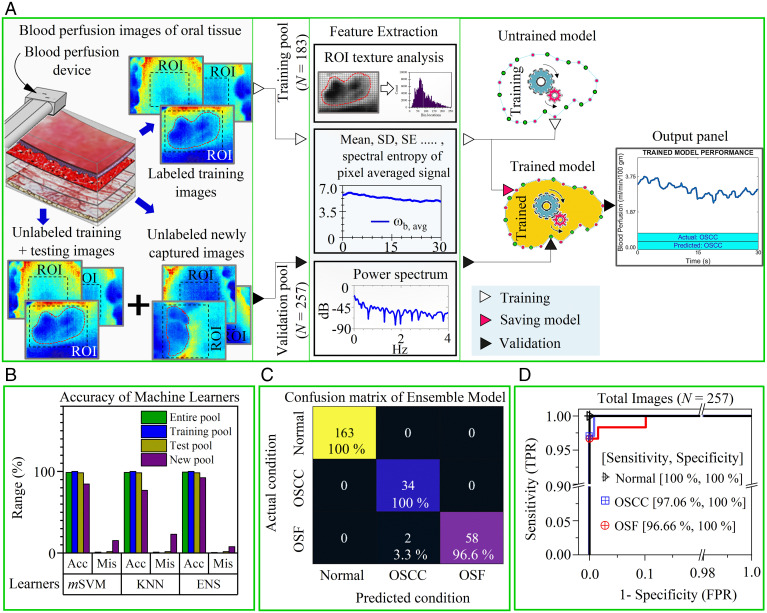
Fig. 5.
ML-enabled OSCC and OSF recognition. (A) Overview of the training and validation process of the ML algorithm to recognize disease condition with high accuracy based on captured blood perfusion images by the device. Here, the training and validation dataset consist of images from all oral sites and the ROI was picked by the operator (or clinician) during acquisition to emphasis on the lesion area. These ROI are subjected to texture analysis and signal processing during feature extraction. The extracted features are passed to an unfitted machine learner with untrained parameters. Subsequently, the fitted model (or trained learner) could be saved and deployed as C or MATLAB scripts to the device processing unit for automatic classification. The model validation is ascertained by passing a pool of randomly distributed unlabeled images (i.e., training, testing, and newly acquired images) through a fitted model. The output console of the processing unit displays the actual and predicted conditions. (B) Comparison of disease detection results (i.e., accuracy [Acc] and misclassification [Mis]) by three different classifiers (mSVM, KNN, and ENS) of testing, the new and the entire image pool. (C) The heat map compares the actual condition (identified by clinical and histological features) and predicted condition identified by best learner (i.e., ensemble model) for the entire data pool (see SI Appendix, Fig. S14 for other pools). (D) ROC curve to represent the performance (i.e., true positive rate [TPR] and false positive rate [FPR]) of the ensemble model to accurately classify one class from other two classes. Areas under the curve for all curves were >0.98.