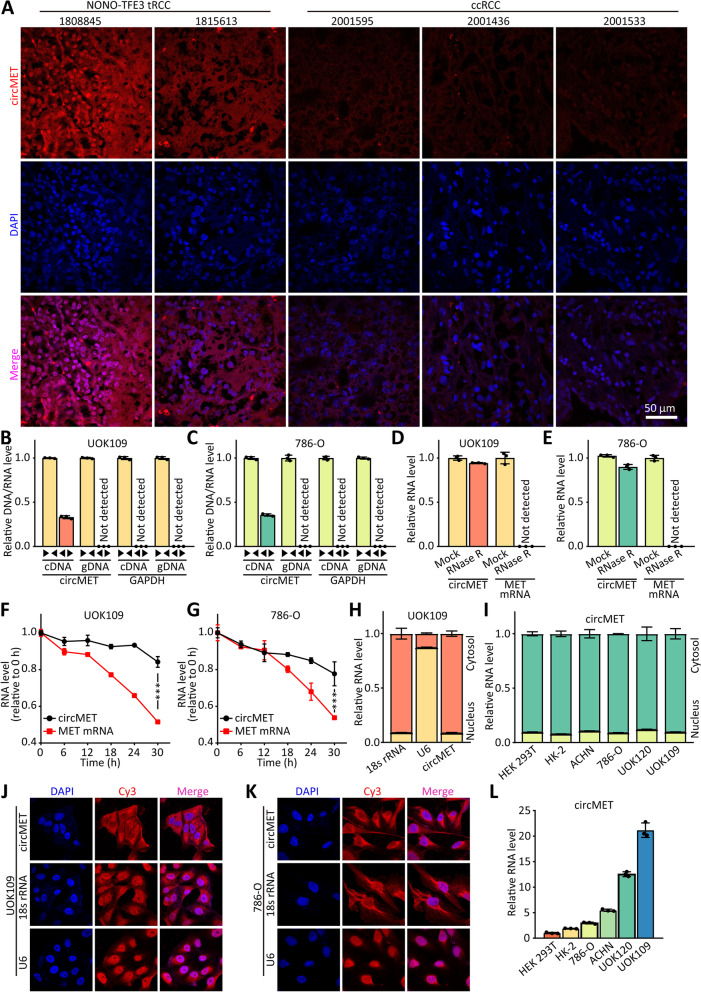
Fig. 1.
CircMET is significantly high-expressed in NONO-TFE3 tRCC. (A) The RNA level of circMET (red) in NONO-TFE3 tRCC and ccRCC was detected by FISH. Nuclei were visualized using DAPI (blue) counterstain. (B-C) cDNA and genomic DNA (gDNA) of UOK109 cells and 786-O cells were used to amplify circMET and GAPDH with divergent primers and convergent primers, respectively. (D-E) The RNA levels of circMET and MET mRNA in both UOK109 cells and 786-O cells were detected by real-time PCR after treatment of RNase R. (F-G) The expression of circMET and MET mRNA in both UOK109 cells and 786-O cells was detected by real-time PCR after treatment with α-amanitin at the indicated time points. (H-I) The subcellular distribution of circMET was analyzed via real-time PCR in ccRCC cell line (786-O and ACHN), tRCC cell lines (UOK109 and UOK120) and normal cell lines (HK-2 and HEK293T). U6 and 18s rRNA were used as nuclear and cytoplasmic markers, respectively. (J-K) The location of circMET (red) in UOK109 and 786-O cells was determined by FISH assay. U6 and 18s rRNA were used as nuclear and cytoplasmic markers, respectively. DAPI-stained nuclei are blue. (L) The RNA level of circMET was analyzed by real-time PCR assay in HEK293T, HK-2, ACHN, 786-O, UOK120 and UOK109 cells. The data are presented as the mean ± SD