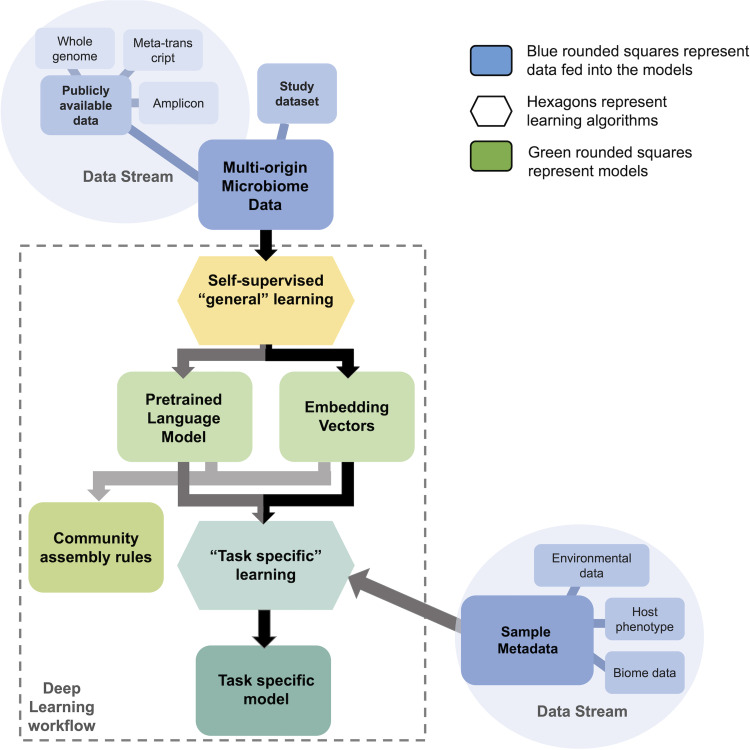
FIG 1.
Workflow of microbial data transformation using deep learning approaches used to capture cross-system properties and apply to a specific task. Hexagons represent algorithms, fed data (in blue) and producing models (squares). Measured microbial features from publicly available data sets are input into the deep learning algorithms (in yellow) to produce pretrained models constituting new features (in green). During pretraining, a model is trained to reconstruct original input from distorted or partial input, using unlabeled data. The process outputs a trained model and/or a set of feature embeddings (which may be interpreted to represent community assembly rules and microbial ecology principles). The general model may then be fine-tuned on metadata (study data set in blue) to answer a biome- or system-specific question of interest. The process of training a model on one task/data set and then fine-tuning it on another task/data set is known as transfer learning.