Figure 7.
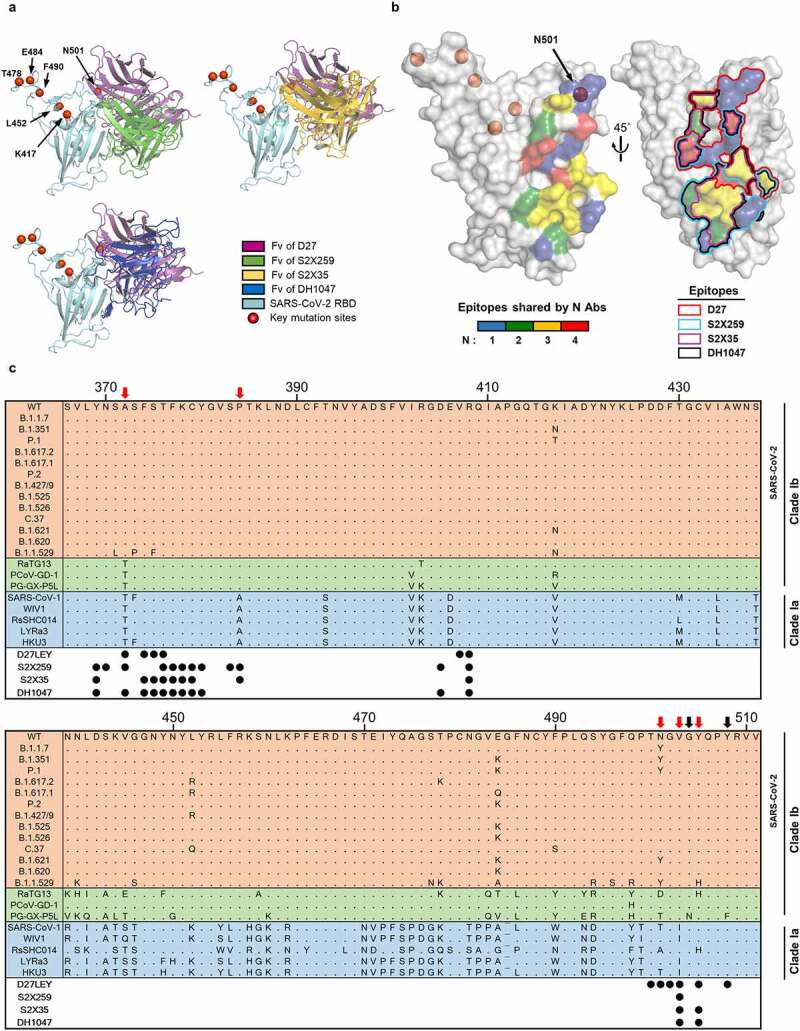
The epitopes recognized by four broadly nAbs. (a) The Fvs of D27, S2X259 (PDB entry: 7RAL), S2X35 (PDB entry: 7R6W) and DH1047 (PDB entry: 7LD1) are shown bound to the RBD (cyan). The red circles indicate key mutation sites shared by many SARS-CoV-2 variants (https://covariants.org/shared-mutations). (b) On the surface of the RBD, the epitopes recognized by the four nAbs are shown, color-coded according to the number of sharing antibodies (Left) and delineated by lines (Right). Epitopes are defined as the RBD residues that contact with ≥ 40 atoms of the bound antibody within 4.5 Å. (c) Alignment of the representative RBD sequences is shown for sarbecovirus clades Ia and Ib, which include the SARS-CoV-2 VOCs, VOIs, and VUMs. The residue positions of the epitopes are indicated by the circles below the alignment. The black arrows indicate epitope residues that are variable in bat and pangolin coronaviruses (RaTG13, PCoV-GD-1, PG-GX-P5L), but not in the SARS-CoV-2 variants. The red arrows indicate epitope residues that are variable across clades Ib and Ia.
