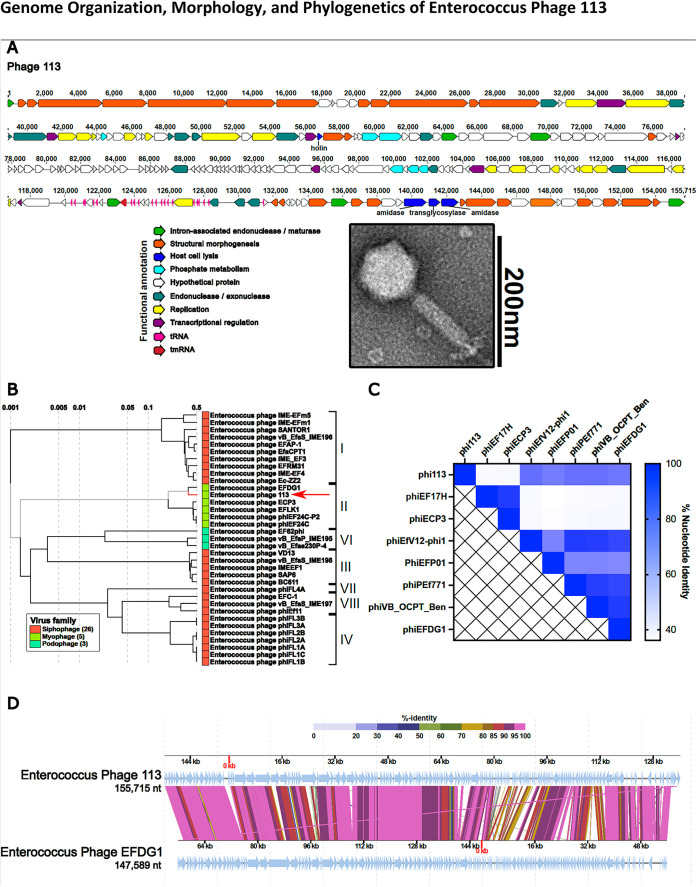
FIG 1.
(A) Whole-genome sequencing reveals a modular organization of phage 113 intermixed with group I introns (bright green). Open reading frames for each phage were determined by the Texas A&M Center for Phage Therapy structural analysis workflow, version 2021.02 (17). Colored open reading frames correspond to functional predictions. The bottom right portion demonstrates the myophage morphotype of phage 113 by TEM. (B) Comparative genome analysis demonstrates that Enterococcus phage 113 clusters with Enterococcus phages of the orthocluster II myophages and most closely resembles phage EFDG1 (19, 20). A proteomic tree was constructed by submitting the Enterococcus phage 113 nucleotide sequence to ViPTree (27). (C) Heat map demonstrating phage 113 nucleotide identity compared to other Enterococcus myophages. The nucleotide identity matrix was determined with progressiveMauve using the Texas A&M Center for Phage Therapy Phage Comparative Genomics workflow, version 2021.01 (17, 28). (D) Enterococcus phages 113 and EFDG1 have high protein sequence homology and similar genome organizations. Colored lines connecting genomes indicate percent protein identity along the length of each genome. The protein-coding sequence alignment was performed using ViPTree (27). nt, nucleotides.