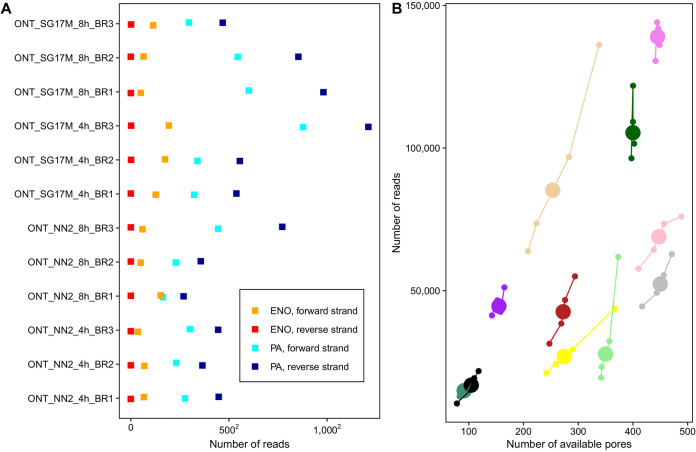
FIG 1.
Evaluation of the Nanopore sequencing runs and genome-based read alignment with minimap2. (A) Representation of the number of P. aeruginosa-specific RNA reads aligning to either the forward (light blue) or the reverse (dark blue) strand of the corresponding reference sequence as well as the number of spike-in RNA control reads mapping to the forward (orange) or reverse (red) human enolase strand. Overall, 4 out of 143,693 total spike-in enolase reads (0.003%) were spuriously antisense associated. Results are shown for the three biological replicates (BR1 to BR3) of SG17M and NN2 at the mid-exponential phase (4 h) and early stationary growth phase (8 h). (B) During Oxford Nanopore sequencing runs, MUX sensor scans of pore fit were performed every 90 min to evaluate the number of available pores for the next 90-min sequencing period. Here, the number of reported available pores after MUX scan and the number of corresponding reads sequenced in the following 90-min period are visualized for the first 6 h. Each flow cell is represented by a unique color. The enlarged circle depicts the group centroid. A strong positive correlation between the number of available pores and the number of sequenced reads was detected (Pearson’s correlation coefficient = 0.8; Pearson’s P value < 0.0001; confidence intervals = 0.73 to 0.86).