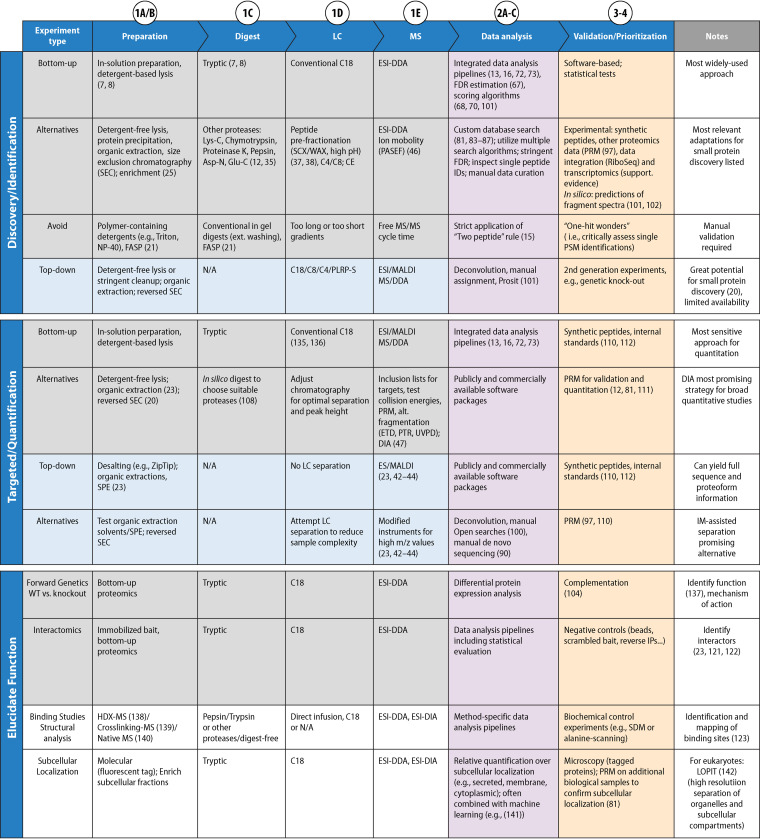
FIG 2.
Overview of the major steps of the most common MS-based workflows for discovery/identification of small proteins, their targeted analysis (for quantification), and for the functional characterization of novel and known small proteins. The numbering of the steps is aligned with Fig. 1, with corresponding text sections indicated by white circles, as follows: 1B, “Sample Preparation and Data Collection—Preparation and enrichment for small proteins”; 1C, “Sample Preparation and Data Collection—Protease digestion”; 1D, “Sample Preparation and Data Collection—Liquid chromatography”; 1E, “Sample Preparation and Data Collection—Ionization and data acquisition”; 2A, “Data Analysis—Overview: the relevance of genome sequences for proteogenomics”; 2B, “Data Analysis—Creation of custom search databases”; 2C, “Data Analysis—Stringent FDR control”; 3, “Validation of Novel Small Protein Candidates”; 4, “Prioritization/Selection of Novel Small Proteins.” Alternative approaches are listed and selected references provided.