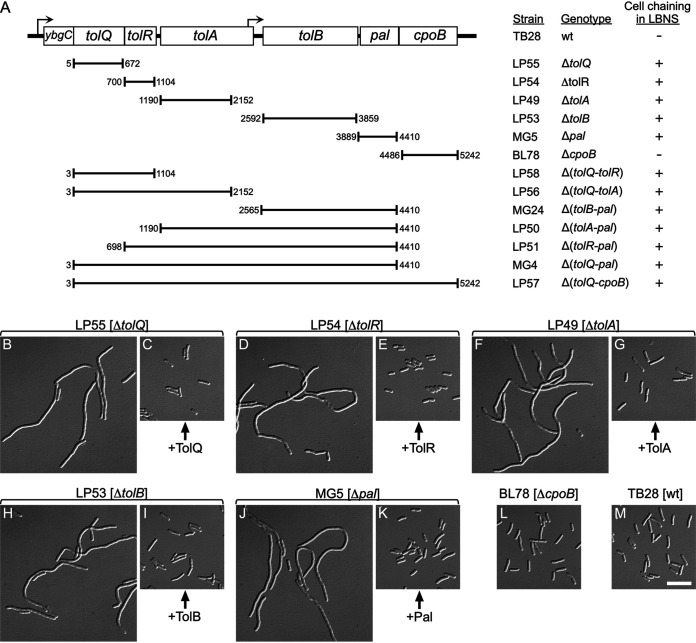
FIG 1.
Strains lacking one or more genes of the tolQ-cpoB cluster. (A) The organization of tol-pal and cpoB genes on the E. coli chromosome is depicted. Hooked arrows indicate the direction of transcription and the locations of predicted promoters (117). Chromosomal fragments that were replaced with aph (kanamycin resistance) to yield the various deletion derivatives of strain TB28 (wt) are indicated below the schematic. Numbers correspond to the first and last base pairs of replaced fragments, counting from the start of tolQ. Columns on the right list corresponding strain names, genotypes, and whether the cells formed chains in LBNS medium (+) or displayed a normal division phenotype (−). (B to M) Cell chaining caused by nonpolar deletions in tolQ, tolA, tolR, tolB, or pal. Differential interference contrast (DIC) images of chemically fixed cells of strains LP55 (ΔtolQ) (B and C), LP54 (ΔtolR) (D and E), LP49 (ΔtolA) (F and G), LP53 (ΔtolB) (H and I), MG5 (Δpal) (J and K), BL78 (ΔcpoB) (L), and TB28 (wt) (M). Strains carried pLP146 (PBAD::tolQ) (C), pLP225 (PBAD::tolR) (E), pCH555 (PBAD::tolA) (G), pMG45 (PBAD::tolB) (I), pCH525 (PBAD::pal) (K), or the vector control pBAD33 (PBAD::) (all others). Cultures were grown to density overnight in regular LB (0.5% NaCl), diluted 200-fold in LBNS (no added NaCl) with 0.05% arabinose, and further incubated for ∼5 mass doublings to optical density at 600 nm (OD600) of 0.6 to 0.7 before fixation. Bar equals 10 μm.