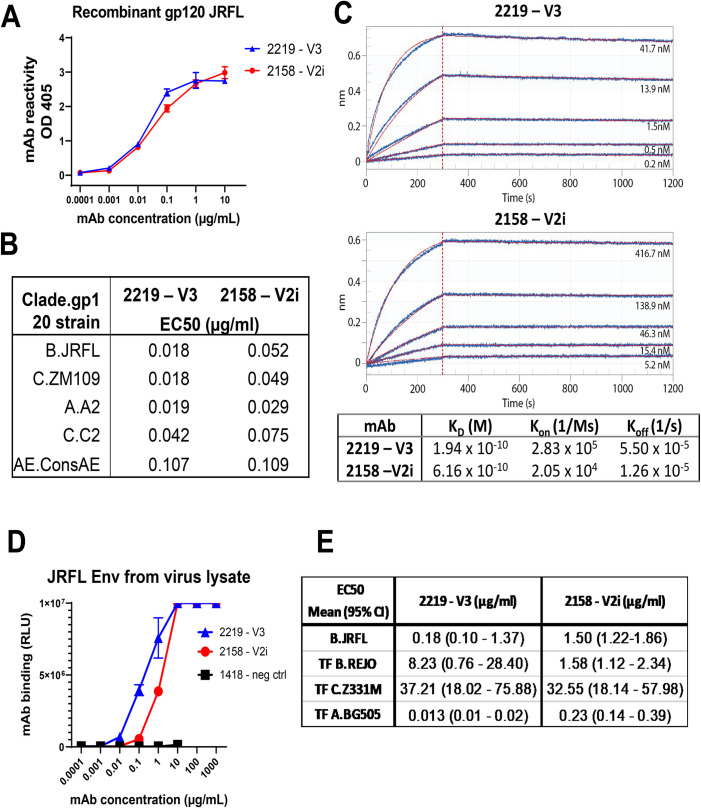
Fig 2. The binding strength of V2i vs V3 mAb to recombinant JRFL gp120 or virus-derived gp120.
A) Direct ELISA measurement of mAb reactivity to recombinant JRFL gp120 (1 μg/ml coated on plates). B) EC50 values of mAb ELISA binding to recombinant gp120 proteins from different HIV-1 strains. C) Kinetics analysis of mAbs binding to recombinant JRFL gp120 by biolayer interferometry. mAbs were captured by anti-hIgG Fc biosensors and reacted with monomeric gp120 in solution for measurement of Fab-gp120 affinity. Fitted curves (1:1 binding model) are shown in red. Molar concentrations of gp120 are indicated. D) Measurement of mAb binding to gp120 from virus-derived solubilized JRFL Env captured with ConA in sandwich ELISA. E) EC50 values (mean and range) of mAb binding to Env derived from JRFL and other HIV-1 strains. HIV-1 clade or CRF is indicated by letters before the strain name. TF: transmitted founder.