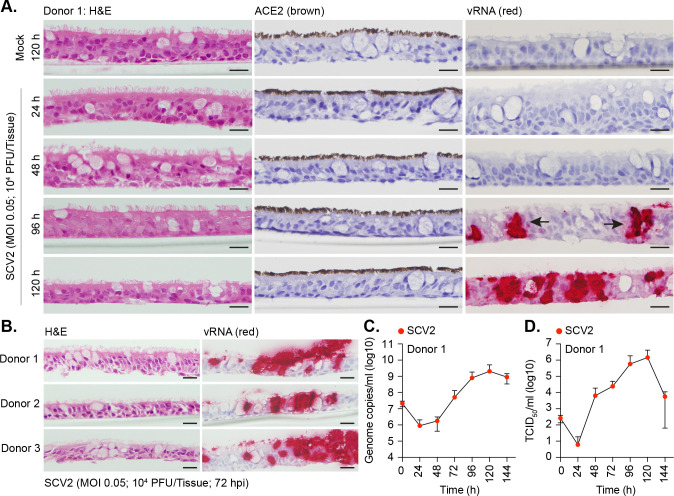
Fig 1. Differentiation of primary bronchial epithelial airway cultures supports SARS-CoV-2 replication.
Primary bronchial epithelial cells isolated from 3 independent donors (donor 1, male Caucasian aged 63 years; donor 2, Hispanic male aged 62 years; donor 3, Caucasian female aged 16 years; all nonsmokers) were seeded onto 6.5 mm transwells and differentiated under ALI conditions for ≥35 days. Ciliated respiratory cultures were mock treated (media only) or SARS-CoV-2 (SCV2; MOI 0.05, 104 PFU/Tissue) infected at 37°C for the indicated times (h). (A, B) Representative images of H&E, ACE2 IHC (brown), or SARS-CoV-2 RNA ISH (red) stained sections. Hematoxylin was used as a counter stain. Scale bars = 20 μm. (C) Genome copies per ml of SARS-CoV-2 in apical washes harvested over time as determined RT-qPCR. N ≥ 7 tissues per condition derived from a minimum of 3 independent biological experiments. Means and SD shown. (D) TCID50 assay measuring infectious viral load in apical washes harvested from SARS-CoV-2 infected tissues over time. Means and SD shown. N = 6 tissues per condition derived from a minimum of 3 independent biological experiments. Raw values presented in S9 Data. ACE2, angiotensin-converting enzyme 2; ALI, air–liquid interface; H&E, hematoxylin and eosin; IHC, immunohistochemistry; ISH, in situ hybridization; PFU, plaque-forming unit; RT-qPCR, reverse transcription quantitative PCR; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; vRNA, viral RNA.