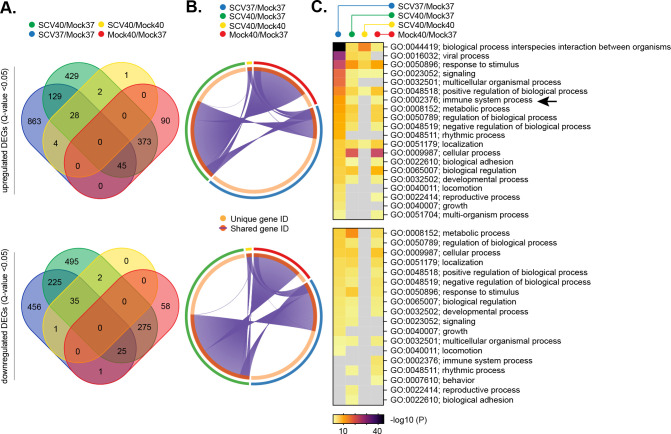
Fig 5. Respiratory airway cultures induce distinct transcriptional host responses to SARS-CoV-2 infection at elevated temperature.
Ciliated respiratory cultures differentiated from primary HBEp cells isolated from 3 independent donors (donor 1, male Caucasian aged 63 years; donor 2, Hispanic male aged 62 years; donor 3, Caucasian female aged 16 years; all nonsmokers) were incubated at 37 or 40°C for 24 h prior to mock (media only) or SARS-CoV-2 (SCV2; MOI 0.05, 104 PFU/Tissue) infection. Tissue were incubated at their respective temperatures for 72 h prior to RNA extraction and RNA-Seq. DEGs (Q < 0.05, up-regulated [top panels] or down-regulated [bottom panels]) were identified for each paired condition analyzed; blue ellipses, SARS-CoV-2 37°C/Mock 37°C (SCV37/Mock37); green ellipses, SARS-CoV-2 40°C/Mock 37°C (SCV40/Mock40); yellow ellipses, SARS-CoV-2 40°C/Mock 40°C (SCV40/Mock37); red ellipses, Mock 40°C/Mock 37°C (Mock40/Mock37). (A) Venn diagram showing the number of unique or shared DEGs between each paired condition analyzed. (B) Circos plot showing the proportion of unique (light orange inner circle) or shared (dark orange inner circle + purple lines) DEGs between each paired condition analyzed. (C) Metascape pathway analysis showing significant DEG enrichment p-value <0.05 (−log10 p-value shown) for each paired condition. Black arrow, immune system process pathway (GO:0002376). Gray boxes, p > 0.05. (A to C) RNA-Seq data derived from RNA isolated from 3 donors (donors 1 to 3) per sample condition derived from 3 independent experiments per donor. Raw values presented in S1, S3 to S6, and S9 Data. DEG, differentially expressed gene; HBEp, human bronchiolar epithelial; PFU, plaque-forming unit; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2.