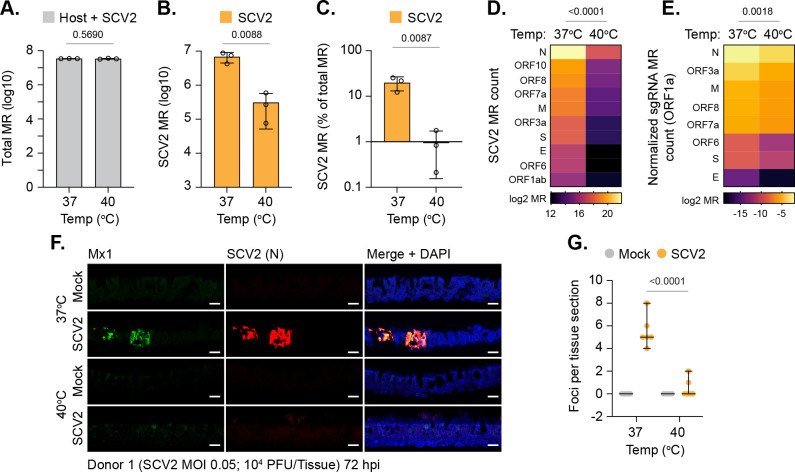
Fig 7. Elevated temperature restricts SARS-CoV-2 transcription in respiratory airway cultures.
Ciliated respiratory cultures differentiated from primary HBEp cells isolated from 3 independent donors (donor 1, male Caucasian aged 63 years; donor 2, Hispanic male aged 62 years; donor 3, Caucasian female aged 16 years; all nonsmokers) were incubated at 37 or 40°C for 24 h prior to mock (media only) or SARS-CoV-2 (SCV2; MOI 0.05, 104 PFU/Tissue) infection. Tissues were incubated at their respective temperatures for 72 h prior to RNA extraction and RNA-Seq. (A) Total MRs (host + SARS-CoV-2) from infected tissues; means and SD shown; p-value shown, unpaired two-tailed t test. (B) SARS-CoV-2 MRs from infected tissues; means and SD shown. (C) % SARS-CoV-2 MRs of total MR count (human + SARS-CoV-2); means and SD shown. (B/C) Unpaired two-tailed t test, p-value shown. (D) Expression values (log2 MR) of SARS-CoV-2 ORFs. (E) Expression values (log2 sgRNA count normalized to ORF1a) of SARS-CoV-2 sgRNAs. (D/E) Paired two-tailed t test, p-values shown. (A to E) RNA-Seq data derived from RNA isolated from 3 donors (donors 1 to 3) per sample condition derived from 3 independent experiments per donor. (F) Indirect immunofluorescence staining of tissue sections (donor 1) showing ISG Mx1 (green) and SARS-CoV-2 nucleocapsid (N, red) expression. Nuclei were stained with DAPI. Scale bars = 20 μm. (G) Quantitation of SARS-CoV-2 N foci in respiratory tissue sections (as in F). N = 6 tissue sections per sample condition. Black line, median; whisker, 95% confidence interval; all data points shown; p-value shown, unpaired two-tailed t test. Raw values presented in S9 Data. HBEp, human bronchiolar epithelial; ISG, IFN-stimulated gene; MR, mapped read; PFU, plaque-forming unit; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; sgRNA, subgenomic RNA.