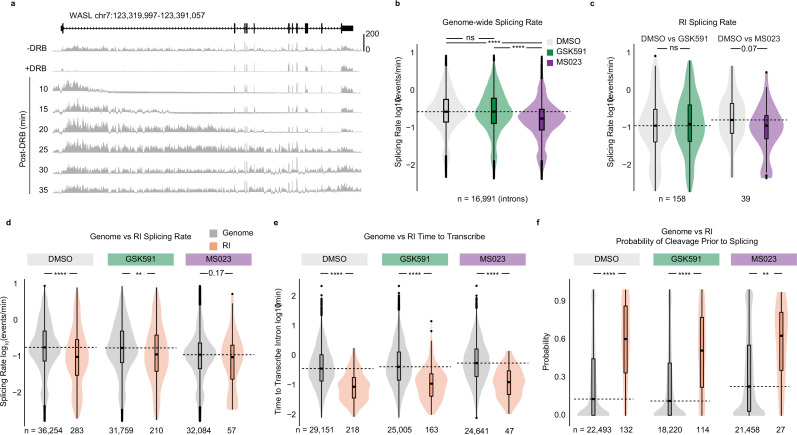
Figure 2. Retained introns (RI) share unique characteristics and are independent of PRMT-regulated co-transcriptional splicing.
(a) Histogram of SKaTER-seq aligned reads across WASL. (b) Distribution of splicing rates for common genomic introns. Dashed line indicates DMSO median; solid line within boxplot is condition-specific median. Significance determined using Wilcoxon rank-sum test; ****<0.0001, ns=not significant. (c) Distribution of splicing rates for common RI as in panel (b). (d–f) Distribution of splicing rates (d), time to transcribe (e), and the probability of cleavage prior to splicing (f) for RI (orange) and A549 expressed introns (dark gray) within the same condition. Dashed line indicates genomic median; solid line within boxplot is condition-specific median. Significance determined using Wilcoxon rank-sum test; *<0.05, **<0.01, ***<0.001, ****<0.0001.