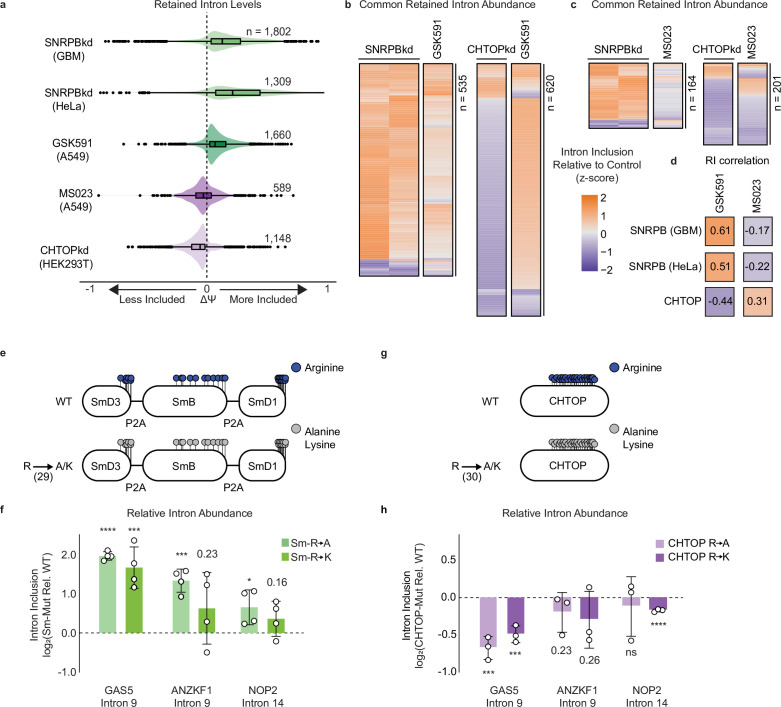
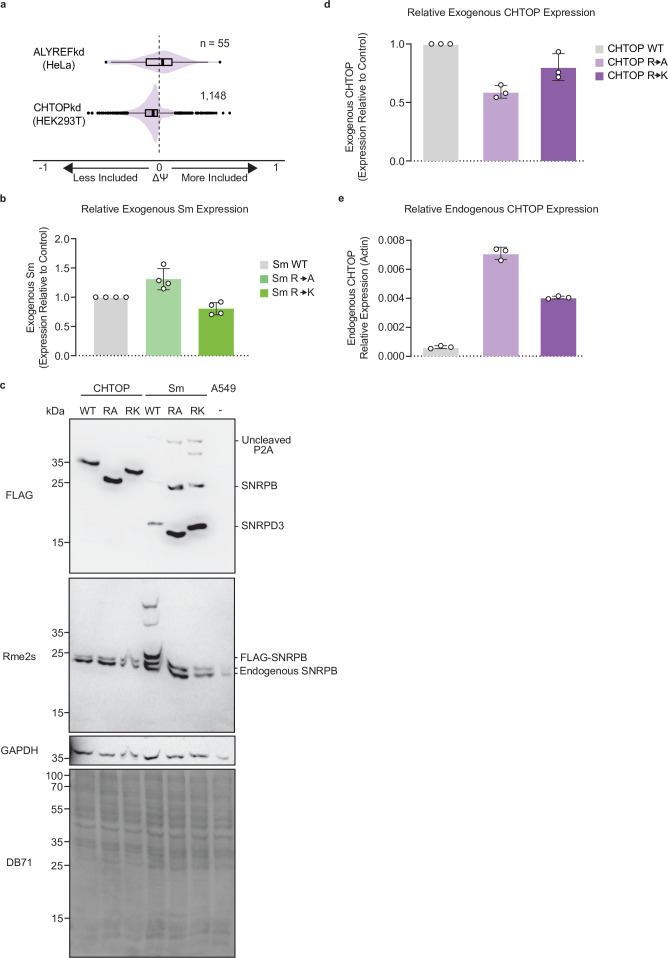
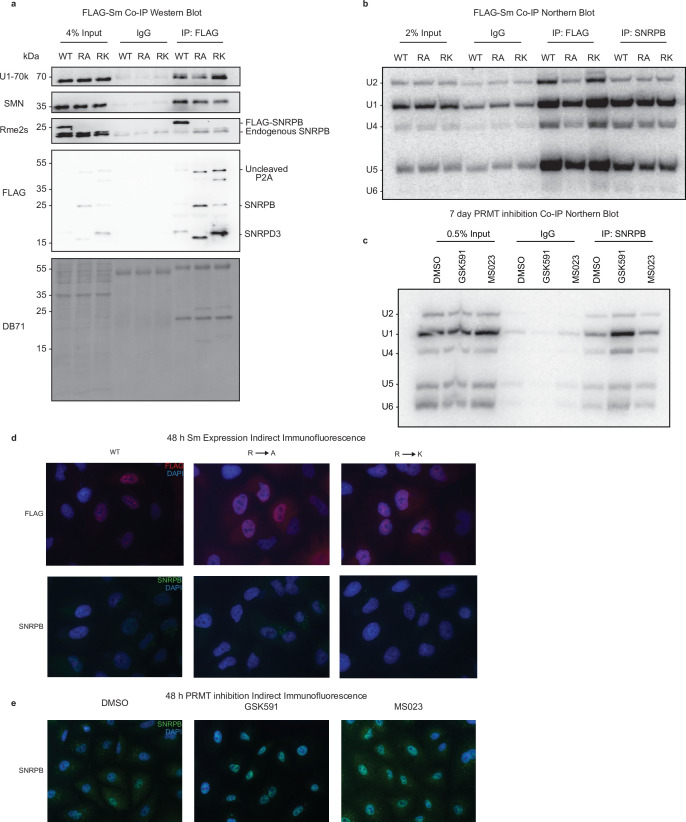
Figure 5. Sm and CHTOP methylarginine mediates PRMT-dependent RI.
(a) Comparison of ΔΨ for RI following SNRPB knockdown (kd), GSK591 treatment, MS023 treatment, and CHTOPkd where ΔΨ=Ψ (Treatment)–Ψ (Control). (b, c) Comparison of ΔΨ z-score for common RI between SNRPBkd and CHTOPkd with either GSK591 (b) or MS023 (c) treatment. (d) Spearman rank correlation of ΔΨ for common RI between SNRPBkd and CHTOPkd with either GSK591 or MS023 treatment. (e) Schematic of Sm expression constructs where lollipops represent individual methylarginines. (f) RT-qPCR of RI following transduction with Sm R-to-A or R-to-K mutants relative to wild-type (WT). Data are represented as mean ± SD. Significance determined using Student’s t-test; *<0.05, ***<0.001, ****<0.0001. (g) Schematic of CHTOP expression construct as in panel (e). (h) RT-qPCR of RI following transduction with CHTOP R-to-A or R-to-K mutants relative to WT. Data are represented as mean ± SD. Significance determined using Student’s t-test; ***<0.001, ****<0.0001, ns=not significant.