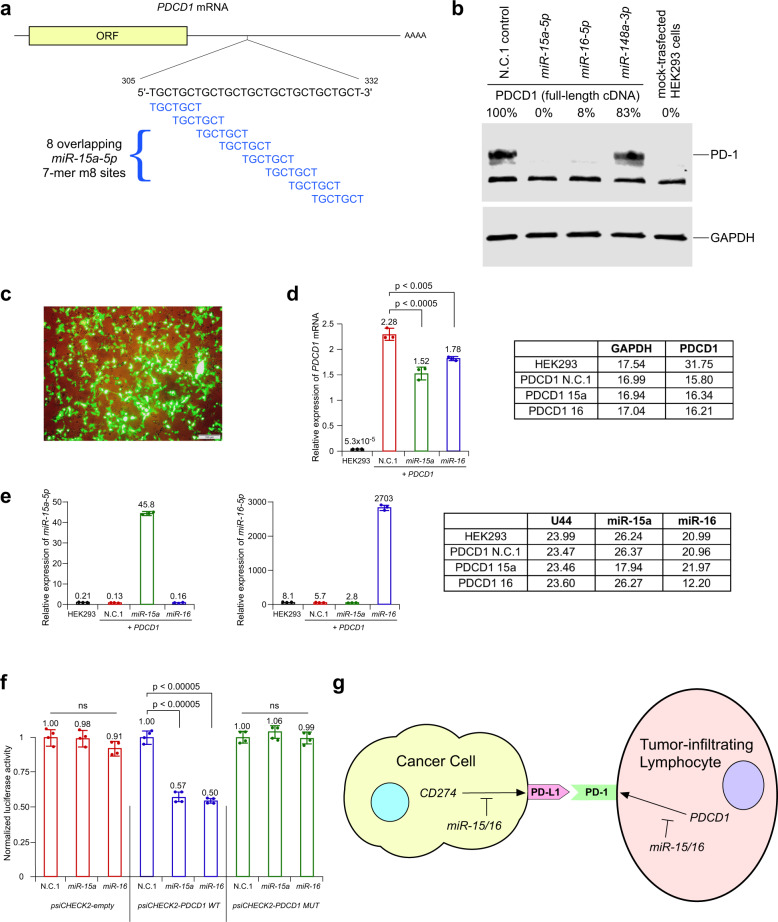
Fig. 1.
miR-15/16 target PDCD1 (PD-1) expression. a A 28 bp DNA fragment containing eight overlapping miR-15/16 target sites in the 3′ UTR of PDCD1. b miR-15/16 inhibit PD-1 protein expression. miR-15a-5p and miR-16-5p expression suppress the PDCD1 expression by targeting its 3’-UTR. Western blot data showed the protein level of overexpressed PDCD1 in HEK293 cells co-transfected with scrambled pre-miR negative control, pre-mir-15a-5p, pre-miR-16-5p and pre-miR-148a-3p mimics. GAPDH served as a loading control. c An example of transformation efficiency in experiments in HEK293 cells for western blot and Luciferase assays (as shown in b, f). d, e Results of real-time PCR experiments on RNAs isolated from HEK293 cells co-transfected with PDCD1 construct and miR mimics (same as in b). d Results of real-time PCR using TaqMan probe for human PDCD1 transcript. Average CT numbers are shown (right). e Results of real-time PCR using TaqMan probes for human miR-15a-5p (left) and miR-16-5p (middle). Average CT numbers are shown (right). f PDCD1 is a direct target of miR-15/16. Renilla luciferase reporter assay showing the reporter expression in HEK293 cells co-transfected with wild-type 3′-UTR of PDCD1 and mutant 3′-UTR of PDCD1 along with scrambled negative control 1, miR-15a and miR-16 mimics. Renilla luciferase activity was normalized to firefly luciferase activity. The normalized luciferase activities in HEK293 cells transfected with different psiCHECK2 constructs and scrambled negative control 1 (pre-miR-N.C.1) were set at 1 and relative luciferase activities of HEK293 cells co-transfected with each psiCHECK2 construct and miR-15a or miR-16 mimics are shown. Two independent experiments were carried out in duplicates and data were presented as mean ± SD. g miR-15/16 are key regulators of PD-1–PD-L1 interaction