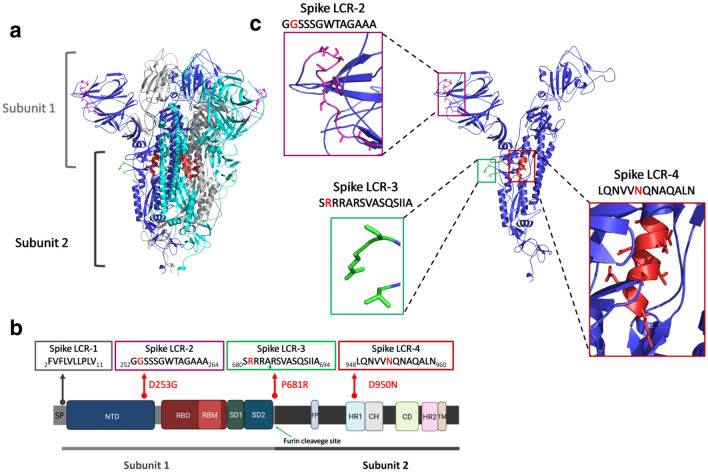
Figure 3.
(a) The SARS-CoV-2 spike protein three-dimensional structure (by cryo-electron microscopy24, PDB code: 7BNM). The structure corresponds to a trimer, where each monomer is represented with a different color. The two subunits that make up each monomer (Subunit 1, also known as Head region, and Subunit 2, or the Stalk region) are indicated. (b) Domain organization of the spike protein. The position of each of the LCRs found in this work, together with the mutations present in each variant spike protein are shown. The position of the signal peptide (SP) and Spike LCR-1 are indicated. The green arrow in the Spike LCR-3 box indicates the furin cleavage site. (c) Monomer of the spike protein. Close ups of each of the structural regions corresponding to the different LCRs are shown in colored boxes. The sequences of each LCR are represented, with the mutations indicated with a red letter. Protein structures in panels a and c were rendered using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). Panel b was created with BioRender.com. Abbreviations: SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain.