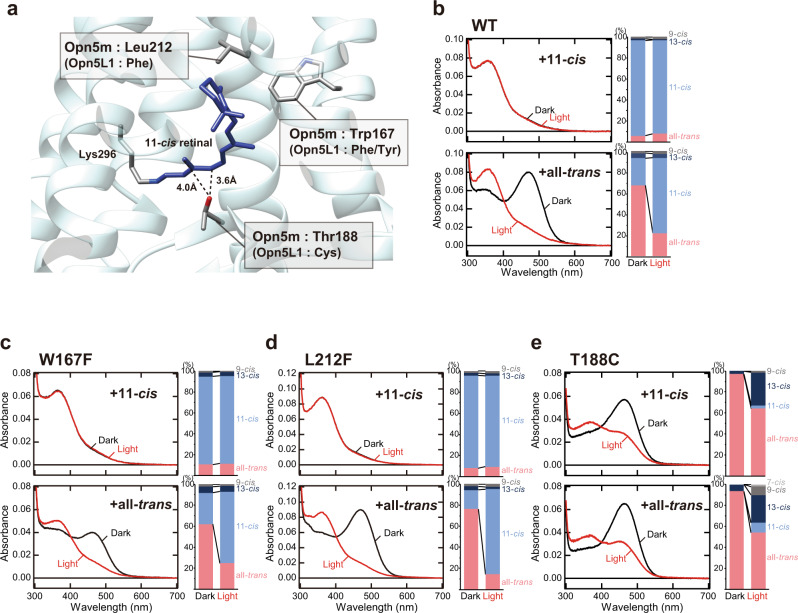
Fig. 1. Binding selectivity of retinal isomers in wild-type and W167F, L212F, and T188C mutant Opn5m proteins.
a Different amino acid residues around the retinal between Opn5m and Opn5L1 subgroups. A structural model was constructed based on the crystal structure of squid rhodopsin (PDB: 2z73) visualized using UCSF Chimera27. Squid rhodopsin has serine and phenylalanine at positions 188 and 212 (based on the bovine rhodopsin numbering system) (Supplementary Fig. 1), which were replaced by threonine and leucine in this model. b-e (left) Absorption spectra of Opn5m wild-type (b) and W167F (c), L212F (d), and T188C (e) mutant proteins purified after the addition of 11-cis or all-trans retinal to the collected cell membranes. The spectra were recorded at 0 °C in the dark (curve 1) and after yellow light (>500 nm) irradiation (curve 2). Spectral change by light irradiation is shown in left panels of Supplementary Fig. 2a–h. (right) Retinal configuration changes of Opn5m wild-type (b) and W167F (c), L212F (d), and T188C (e) mutant proteins purified after the addition of 11-cis or all-trans retinal to the collected cell membranes. The retinal isomers before and after yellow light (>500 nm) irradiation were analyzed with HPLC after extraction of the chromophore as retinal oximes (right panels of Supplementary Fig. 2a–h). The negative peak in the visible region of the difference spectrum calculated before and after yellow light irradiation of the wild-type (Supplementary Fig. 2e) corresponds to λmax of the all-trans retinal bound form (474 nm). λmax of the all-trans retinal bound form of the mutant proteins calculated from their difference spectra (Supplementary Fig. 2f–h) are 470 nm (W167F), 475 nm (L212F), and 470 nm (T188C).