Fig. 2.
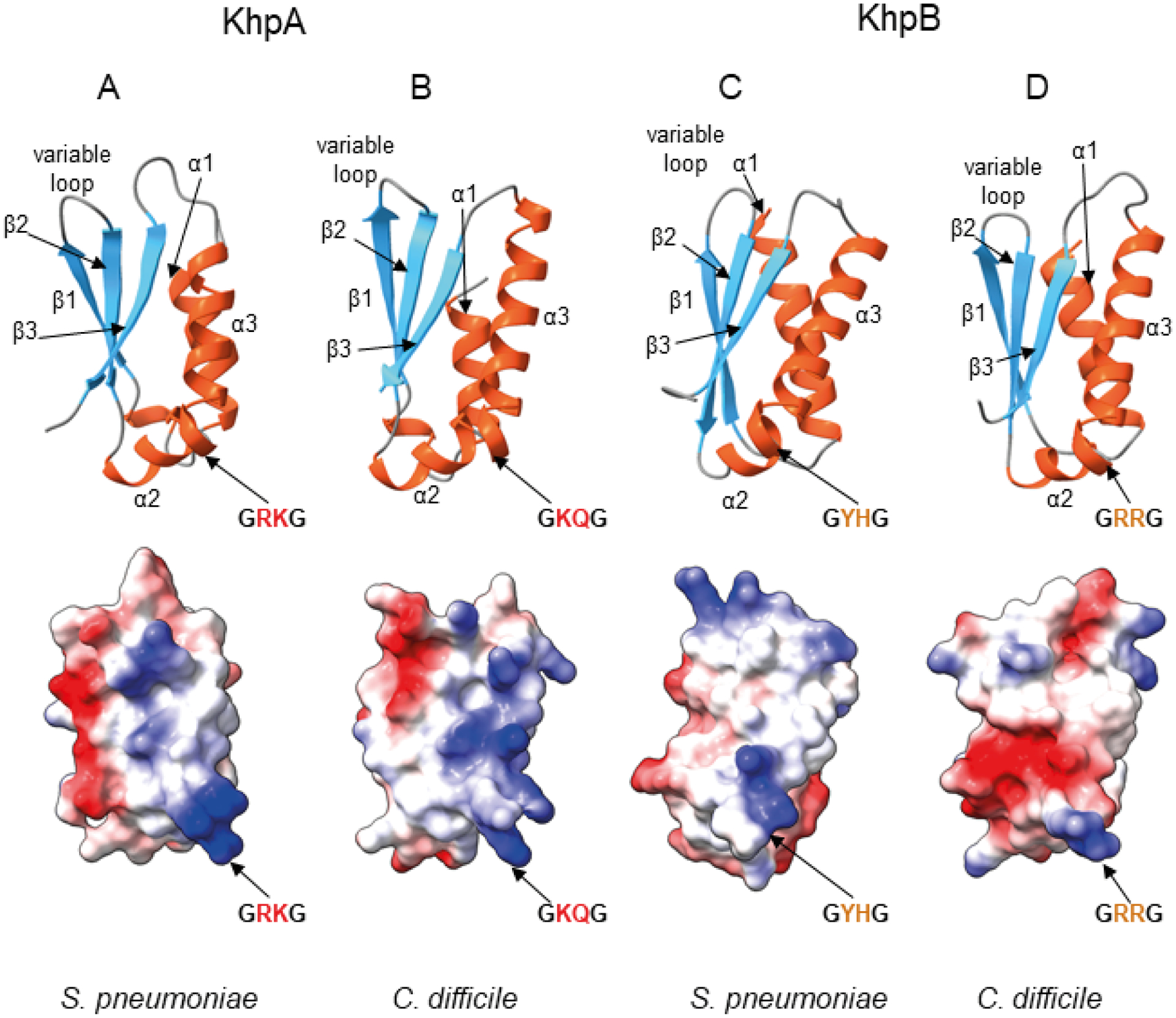
Predicted structures of the KH domains of KhpA and KhpB proteins from S. pneumoniae and C. difficile reveal the same overall fold but different electrostatic surface potential.
A. Predicted structure of KhpA from S. pneumoniae.
B. Predicted structure of KhpA from C. difficile.
C. Predicted structure of KH domain of KhpB protein from S. pneumoniae.
D. Predicted structure of KH domain of KhpB protein from C. difficile. All structures were predicted using ColabFold software (Mirdita et al., 2021) based on AlphaFold 2.0 (Jumper et al., 2021) and MMseqs2 (Steinegger & Soding, 2017), and visualized using ChimeraX (Pettersen et al., 2021). In each pair, a ribbon representation is shown above with α-helices shown in red, and β-strands in blue, and an electrostatic surface potential, calculated using ChimeraX, is shown below. The view of the KH domains is at the face involved in RNA binding.
