Fig. 3.
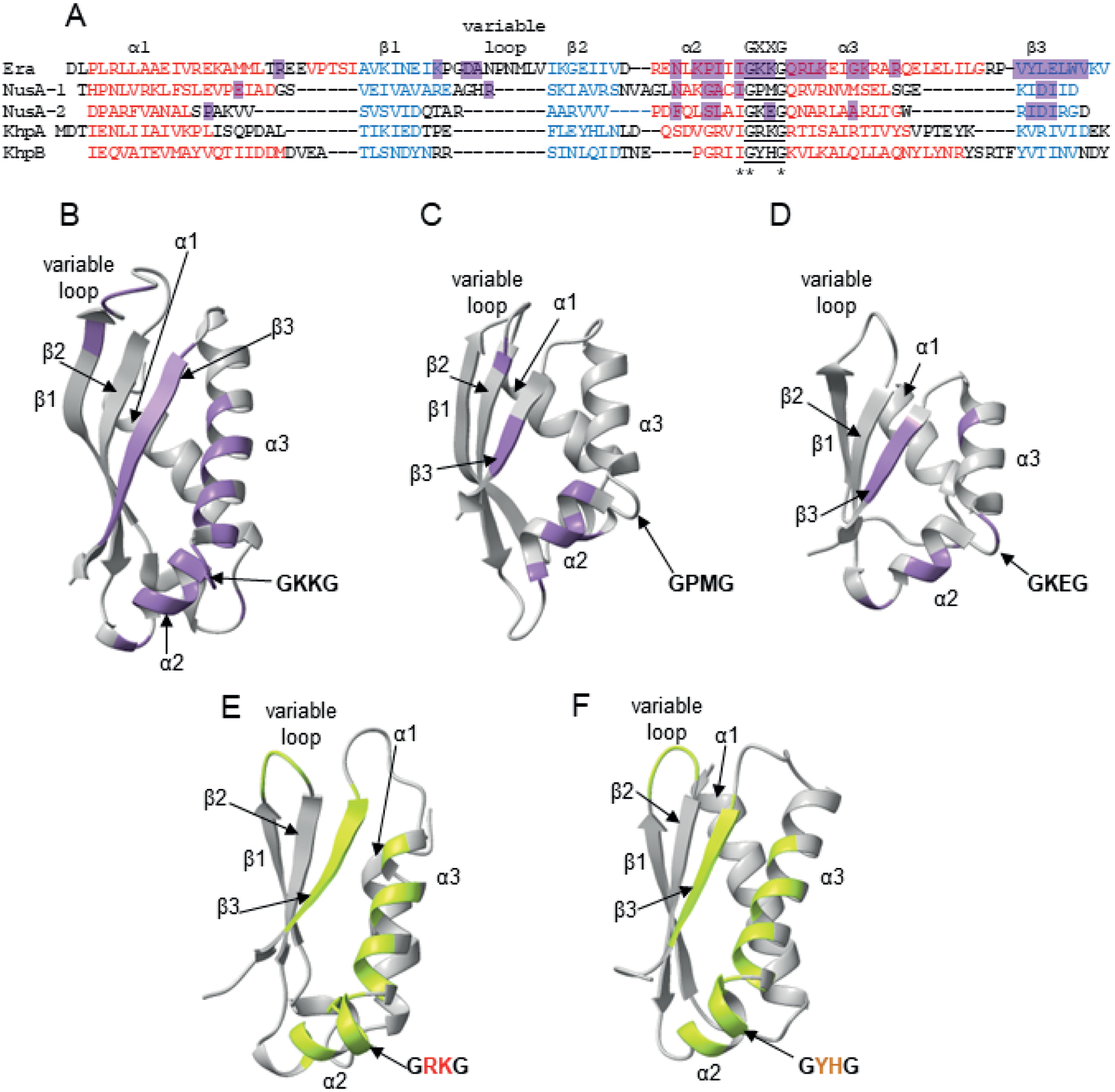
Structurally-determined and predicted RNA binding contacts in KH domains.
A. Alignment of the structurally homologous regions of the KH domain from A. aeolicus Era protein (Tu et al., 2009), the KH domain 1 from M. tuberculosis NusA protein, the KH domain 2 from M. tuberculosis NusA protein (Beuth et al., 2005), and KH domains from Alphafold-predicted S. pneumoniae KhpA and KhpB. For the alignment, homologous sequences were first aligned using Clustal Omega, and then structurally homologous regions were manually aligned based on the Era and NusA structures (Beuth et al., 2005, Tu et al., 2009) and the predicted structures of KhpA and KhpB.
B. The structure of the KH domain of A. aeolicus Era with RNA contacts according to (Tu et al., 2009) marked purple.
C. The structure of the KH 1 domain of M. tuberculosis NusA with RNA contacts according to (Beuth et al., 2005) marked purple.
D. The structure of the KH 2 domain of M. tuberculosis NusA with RNA contacts according to (Beuth et al., 2005) marked purple.
E. The predicted structure of full-length S. pneumoniae KhpA, in which regions that could hypothetically be involved in RNA binding are marked lime-green.
F. The predicted structure of the KH domain of S. pneumoniae KhpB, in which regions that could hypothetically be involved in RNA binding are marked lime-green.
The structures of S. pneumoniae KhpA and KhpB proteins were predicted using ColabFold (Mirdita et al., 2021), and all structures were visualized using Chimera X (Pettersen et al., 2021).
