Abstract
Objective:
Quantify tradeoffs in performance, reproducibility, and resource demands across several strategies for developing clinically relevant word embeddings.
Materials and Methods:
We trained separate embeddings on all full-text manuscripts in the Pubmed Central (PMC) Open Access subset, case reports therein, the English Wikipedia corpus, the Medical Information Mart for Intensive Care (MIMIC) III dataset, and all notes in the University of Pennsylvania Health System (UPHS) electronic health record. We tested embeddings in six clinically relevant tasks including mortality prediction and de-identification, and assessed performance using the scaled Brier score (SBS) and the proportion of notes successfully de-identified, respectively.
Results:
Embeddings from UPHS notes best predicted mortality (SBS 0.30, 95% CI 0.15 to 0.45) while Wikipedia embeddings performed worst (SBS 0.12, 95% CI −0.05 to 0.28). Wikipedia embeddings most consistently (78% of notes) and the full PMC corpus embeddings least consistently (48%) de-identified notes. Across all six tasks, the full PMC corpus demonstrated the most consistent performance, and the Wikipedia corpus the least. Corpus size ranged from 49 million tokens (PMC case reports) to 10 billion (UPHS).
Discussion:
Embeddings trained on published case reports performed as least as well as embeddings trained on other corpora in most tasks, and clinical corpora consistently outperformed non-clinical corpora. No single corpus produced a strictly dominant set of embeddings across all tasks and so the optimal training corpus depends on intended use.
Conclusion:
Embeddings trained on published case reports performed comparably on most clinical tasks to embeddings trained on larger corpora. Open access corpora allow training of clinically relevant, effective, and reproducible embeddings.
Keywords: word embeddings, clinical informatics, natural language processing, protected health information
Graphical abstract
BACKGROUND AND SIGNIFICANCE
Word embeddings provide a means of converting words and phrases into arrays of numbers so that they can be used in models requiring numerical input. Embeddings are developed through unsupervised learning procedures that aim to capture the semantic meaning and relationships of language in the text. An efficient embedding is able to represent these relationships in a lower-dimensional space than could be accomplished with count-based methods. While such embeddings are useful across many domains, their use in analyzing the text of clinical encounter notes presents several challenges. First, while there are many publicly available pretrained embeddings, many of these are derived from a broad range of non-medical text sources and thus may not capture the appropriate word sense, linguistic relations, or vocabulary needed in a clinical context.[1] Second, while it may be appealing to train new embeddings on locally available clinical notes within a health system,[2] the size of available clinical corpora may vary by center and may not be sufficient to train effective embeddings.[3] Additionally, locally sourced embeddings, even when cleared of personally identifiable information (PII), can be attacked to expose patient-level protected health information (PHI).[4]
Prior work on domain-specific clinical word embeddings has leveraged multiple data sources in addition to text,[5] focused on knowledge discovery in large corpora of biomedical text,[1,2,6-8] utilized the entire[2,8] or only medically-relevant articles on Wikipedia,[9] MEDLINE abstracts,[10] and leveraged the Medical Information Mart for Intensive Care III (MIMIC-III) dataset which is de-identified, but requires a data use agreement for researchers and is limited to a single center.[11] Other work has focused on concept embeddings that capture clinical information from both structured and unstructured data sources and relies on pre-processing with the Unified Medical Language System (UMLS) thesaurus.[12] Studies have compared word embeddings built using different training corpora and training algorithms[13] with mixed results for publicly available corpora compared to local clinical text for training,[1,2] size of training corpus,[1,6] vector dimension,[7] and training algorithm.[2,6,7] For example, BioWordVec is a downloadable embedding set trained on all Pubmed Central (PMC) titles and abstracts; however, its only clinical validation was in semantic similarity and it was not compared to embeddings trained on local clinical text.[14] However, the optimal approach to producing word embeddings that are relevant for the analysis of clinical text, are efficient to train and use, and are free of PII, and thus fully shareable, remains unknown. Additionally, the performance tradeoffs in using corpora that are publicly available and free of PII compared to locally available corpora with PII are unknown.
To overcome these knowledge gaps, we propose published clinical case reports (already fully de-identified as a result of publication, comprises a corpus much smaller than the general corpus of all published manuscripts, not all of which may not be relevant for clinical notes, is readily available for download, and are written in a style analogous to the text of clinical encounter notes) as a text corpus for training word embeddings for clinical natural language processing (NLP) tasks. In this study, we evaluated 60 word embedding sets and compared performance across embedding size, training method, and training corpus. We focused evaluation on previously developed intrinsic and extrinsic NLP tasks relevant to clinical text including linguistic regularity, mortality prediction, de-identification[15], and others. We hypothesized 1) that embeddings trained on the full text of published clinical case reports would share similar lexical and syntactic properties with clinical notes; 2) that such clinically relevant embeddings would demonstrate comparable performance to embeddings trained on corpora of clinical notes that include PHI; 3) that embeddings trained on case reports would outperform embeddings derived from non-medical domain text and general, non-clinical scientific text; and finally 4) that subword n-grams[16] would outperform word-level n-grams[17] due to their ability to produce word vectors for out-of-vocabulary and misspelled terms, which occur frequently in electronic health record (EHR) data.[18]. Finally, we make our embeddings available for public download to support reproducibility and transparency.
MATERIALS AND METHODS
We trained a set of word embeddings using every combination of three training algorithms, five text corpora, and four dimension sizes, for a total of 60 sets of word embeddings (Table 1).
Table 1:
Summary of training algorithms, text corpora, and vector dimensions used for training word embeddings. For training algorithm, word-level describes training on whole tokens in the training corpus while sub-word describes training on n-grams.
| Training Algorithm | Text Corpus | Vector Dimension |
|---|---|---|
| word2vec (word-level) | MIMIC-III (MIMIC) | 100 |
| fasttext (sub-word) | PMC Open Access Subset- All manuscripts (OA-All) | 300 |
| GLoVE (word-level) | PMC Open Access Subset- Case reports only (OA-CR) | 600 |
| University of Pennsylvania Health System (UPHS) | 1200 | |
| Wikipedia- English (Wiki) |
Text Corpora
PMC Open Access Subset
The PMC Open Access Subset- Case reports only corpus (OA-CR) was built using case report manuscripts downloaded from PubMed Central published from 2007-2017 that are available under the OpenAccess Subset. Of the 515,592 reports indexed in the PMC (Supplemental Methods), 27,575 had the full text openly available. Text from both the abstract and manuscript body sections of downloaded XML files were included in the final training corpus. All non-English text was removed using the detect method of the langdetect[19] python package. All text was converted to lowercase and stripped of non-body text, XML tags, break and tabs, figure tables and captions, figure references, citations, and URLs. Reports with less than 100 tokens of processed text were removed.
The PMC Open Access Subset- All manuscripts corpus (OA-All) was built using all manuscripts downloaded from PubMed Central published from 2007-2017 that are available under the OpenAccess Subset. Of the 5,834,856 reports indexed in the PMC (Supplemental Methods), 630,885 had their full text openly available.
MIMIC-III
The MIMIC corpus was built using patient encounter notes from the MIMIC-III dataset.[20,21] 278,269 notes were extracted and processed. All text was converted to lowercase; stripped of end fields, generic fields, de-identification metastrings, underscores used as separation lines, and breaks and tabs; and all non-English text was removed. Clinical notes were often short but appeared valid on manual review, and so a 50 token cutoff was used for inclusion in the final corpus.
Wikipedia
The Wikipedia-English corpus (Wiki) was built using a Wikipedia dump (downloaded 2018-11-02, 15.6GB). The download contained 18,906,413 articles that were processed using the WikiCorpus from the genism python package[22] to provide a standardized and reproducible workflow. Similarly to MIMIC-III notes, a 50 token cutoff was used for inclusion in the final corpus.
University of Pennsylvania Health System
From the University of Pennsylvania Health System (UPHS), we retrieved all signed clinical encounter notes from inpatient and outpatient encounters from January 1, 2017 to December 4, 2019. This corpus contained 14,828,230 notes. Raw text was processed identically to the MIMIC corpus and no de-identification was performed prior to training the embedding models.
Text Processing
Multi-word expressions
We constructed a multi-word expression dictionary of terms to capture relevant meaning of clinical concepts across corpora. Multi-word expressions (MWEs) were identified using both an intrinsic and extrinsic method. First, pointwise mutual information (PMI) was calculated for every bigram and trigram in the OA-CR corpus using the NLTK python package.[23] Bigrams and trigrams were included if they appeared in at least 10 unique manuscripts and were in the 50th to 95th percentile of PMI (percentile based on manual review to determine clinical relevance). Second, the National Library of Medicine specialist lexicon[24] was used to identify MWEs. Any term in the specialist lexicon present in the OA-CR corpus was considered a MWE. In total, 398,217 n-grams were classified as MWEs; a representative sampling of included and excluded MWEs is included in the supplemental methods (Supplemental Tables 1-2). The flashtext python package[25] was used to join all MWEs with an underscore between words in all corpora used for training word embeddings and all text used in evaluation tasks.
Tokenization
Text corpora were processed for training as follows: the OA-CR, MIMIC, and UPHS corpora were first tokenized by sentence and then by word using the spaCy tokenizer in python[26]. Due to the their larger size, the OA-All and Wiki corpora were tokenized using the gensim LineSentence method. Corpora were saved as single text files for GLoVE training.
Word embeddings
Training Algorithms
Word embeddings were trained using word2vec[17] and fasttext[16] with a skip-gram architecture as implemented in the python gensim package.[22] We also trained GLoVE embeddings using the GLoVE C implementation from Stanford NLP (version 1.2).[27]
Tasks
To compare the embedding training procedures, all 60 embeddings were tested in every designed task, irrespective of the source dataset for the task. Intrinsic evaluation tasks were chosen to evaluate the ability of the embeddings to capture semantic representations in a clinical context. Extrinsic tasks were chosen to test the different embeddings in downstream NLP tasks that researchers or data scientists may encounter in working with clinical text.
Intrinsic Evaluation
Semantic Similarity
Semantic similarity was measured by computing the correlation between the cosine similarity of word pairs and the manually curated similarity scores of those pairs in the University of Minnesota Semantic Relatedness Standard (UMNSRS) similarity dataset.[28] This task measures the degree to which an embedding's numeric representation of two terms is as similar as a clinician's assessment of their similarity in a clinical context. Correlations are reported using Spearman’s ρ with 95% confidence intervals. Only word pairs for which both terms had vector representation in the word embedding model were considered in the comparison. For example 'Famvir' and 'bedwetting' were not present in the OA-CR word2vec embedding vocabularies. The percent of UMNSRS terms that were out-of-vocabulary for each training algorithm-corpus pair, as well as the full list of OA-CR word2vec out-of-vocabulary terms are reported in Supplemental Tables 3 and 4 respectively.
Linguistic Regularity
A known feature of continuous space language models is the preservation of an offset vector that captures some semantic regularity. For example, in non-clinical settings, embeddings have maintained semantic relationships such as singular/plural and male/female through an offset vector.[29] By extension, a useful set of clinically relevant word embeddings should capture offset vectors related to medical treatment. We curated a list of 100 pairs of medical terms with a relationship "is_a_treatment_for' across inpatient, outpatient, medical, and surgical contexts likely to be discussed in a clinical encounter note (Supplemental Table 5). For example metformin is a treatment for diabetes mellitus just as lisinopril is a treatment for hypertension. Therefore, an embedding that captures clinically relevant semantic information should reproduce the analogy. νmet formin−νdiabetes = νlisinopril−νhypertension.
To determine whether this treatment relationship was preserved in the vector space for each embedding we used two approaches. First, we calculated the offset as the difference for each term pair from the average cosine similarity across all pairs. We also calculated the offset vector from the cosine similarity of the centroid of disease terms to the centroid of treatment terms (Supplemental Figure 2) and the standard deviation of all pairs to the centroid cosine similarity (Supplemental Figure 4). The offset vector likely does not represent an actual word in the embedding space, rather, it is the regularity identified in training the embedding space. The standard deviation of the cosine similarity for each term pair and the centroid are reported as measures of the regularity of this clinical relationship.
Second, we evaluated linguistic regularity using a previously reported analogy completion task.[30] For every combination of pairs, representing an analogy a:b::c:d, we calculated the cosine similarity between d and the single closest vocabulary word in the embedding space to a−b + c. We report the mean and standard deviation of the cosine similarity across analogies derived from the 100 pairs of medical terms with a relationship "is_a_treatment_for" (Supplemental Figure 5).
Extrinsic Evaluation
Lexicographic Coverage
We evaluated embeddings on their ability to provide a vector representation for all words in a clinical note. A useful set of word embeddings should be able to produce representations for a wide variety of medical terms, including for new terms that might not have been seen in an original training corpus and for misspellings. We tested coverage on 362,430 unique words in the 53,425 MIMIC-III discharge summary notes[20,21] and 398,662 unique words in 48,432 UPHS ICU discharge summary notes. Coverage is reported as the proportion of all unique tokens for which a model could provide a word vector.
Clustering Purity
We evaluated word embeddings for their ability to cluster similar notes using two different sets of discharge summaries from the MIMIC-III and UPHS datasets. The clustering task was performed independently on each set.
Summaries from three ICUs in the MIMIC-III dataset: neonatal intensive care unit (‘NICU’), trauma surgical intensive care unit (‘TSICU’), and coronary care unit (‘CCU), and from four matched ICUs in the UPHS dataset: neonatal intensive care unit (‘NICU’), trauma surgical intensive care unit (‘TSICU’), medical intensive care unit (‘MICU’), and heart and vascular intensive care unit (‘HVICU’) were used for independent clustering experiments. We limited the analysis to a subset of ICUs that are clinically distinct to simplify the clustering task in order to best evaluate the underlying embeddings.
Each discharge summary was given a document-level vector representation by taking the centroid across all word vectors in the note. Out-of-vocabulary words were ignored. A k-means (with k equal to the number of ICU labels in the testing dataset) procedure was used to cluster the document vectors. To evaluate the overall ability of the word embedding representation of discharge summary text, we use the summary statistic clustering purity. Clustering purity was calculated using the formula: where N = total number of test notes, k = number of clusters, C = set of clusters, ci = cluster i in C, and tj = ICU label j. We reported the 95% confidence interval over 1,000 bootstrapped replicates for the purity measure.
Mortality Prediction
We evaluated word embeddings for their ability to capture clinically relevant signal in mortality prediction. Mortality prediction was performed using notes from the MIMIC-III and UPHS datasets. The first physician encounter note charted within 24 hours of hospital admission was used to predict in-hospital mortality during the current admission using a convolutional neural network with a previously described architecture[31] designed for word embedding based input (Supplemental Figure 2). The text of each note was converted into sentence vectors by taking the centroid of a word vector matrix, where every row in the matrix is a vector representation of a word in that sentence. Out-of-vocabulary tokens were ignored. Notes were limited to the first 100 sentences and notes shorter than 100 sentences were padded using sentences from the beginning of the note (see Supplemental Table 6). An 80/20 split was used for training and testing, and sampling was stratified to maintain balance in outcome between the two sets. We reported predictive performance for each model with the scaled Brier score (SBS)[32] and 95% bootstrapped confidence interval over 1,000 replicates. The UPHS dataset was downsampled to reflect the same mortality event rate as the MIMIC dataset to facilitate comparisons.
De-identification
De-identification of clinical encounter notes using word embeddings was performed according to a previously published method.[15] For every token in a note, a vector was randomly sampled for the N nearest word vectors, where N varied between 3,5, and 7 to increase obfuscation. We evaluated embeddings trained on a published corpus lacking PII (OA-AII), one local corpus containing PII (UPHS), and one general corpus where PII removal is not relevant (Wiki). Additionally, we limited the analysis to a single training algorithm (word2vec) and dimension (300) to isolate the corpus as the testing variable.
To assess the quality of the de-identification, 50 de-identified notes from the UPHS corpus were chosen randomly for qualitative review by a physician (GEW). Notes were judged as fully de-identified or not based on obfuscation of both the patient's name and age in the note. Cases where "david" was replaced with "david," or "joan" was replaced with "joans" were not considered sufficient for de-identification. We reported the absolute number and proportion of notes with adequate de-identification. Additionally, once notes were de-identified, we repeated the mortality prediction task as described above on the UPHS notes to assess the clinical relevance of notes obfuscated using the embedding based obfuscation procedure. As a secondary analysis, we also repeated the de-identification procedure using the same three embeddings with 8 notes from the 2014 i2b2 de-identification task and manually evaluated all fields labeled as containing PII.[33]
Ranking
Summary performance of models was compared across the three training variable categories: algorithm, corpus, and vector dimension. To determine ranking for a specific task, mean performance for a category (e.g. training algorithm) was calculated over the remaining categories (e.g. text corpus and vector dimension). The variables in a category were then ranked from best performing (1) to lowest (number of variables in the category, e.g. 3 for training algorithm). De-identification was excluded from this composite evaluation as not all corpora, training algorithms, and vector dimension sizes were compared.
RESULTS
Training corpora varied widely in the number of available documents and tokens (Table 2, Supplemental Table 7). The word2vec and fasttext models identified the same relevant vocabulary from each corpus while the GLoVE embeddings contained a larger vocabulary.
Table 2:
Summary of word embeddings by text corpus. Vocabulary size indicates the number of words that have vector representation in the embedding sets for each training corpus. Abbreviations: PII = patient identifiable information, DUA = data use agreement, OA-CR = PMC Open Access Subset- Case reports only, MIMIC = MIMIC-III clinical notes, OA-All = PMC Open Access Subset- All manuscripts, Wiki = Wikipedia-English, UPHS = University of Pennsylvania Health System clinical encounter notes.
| Corpus | PII | Public Availability |
Documents | Tokens | Vocabulary size- word2vec |
Vocabulary size- fasttext |
Vocabulary size- GLoVE |
|---|---|---|---|---|---|---|---|
| OA-CR | No | Creative Commons License | 27,449 | 49,590,835 | 333,360 | 333,360 | 435,835 |
| MIMIC | No | With approval and DUA | 220,453 | 148,089,760 | 160,411 | 160,411 | 267,629 |
| OA-All | No | Creative Commons License | 628,404 | 1,848,856,520 | 3,748,342 | 3,748,342 | 3,755,370 |
| Wiki | No | Creative Commons License | 4,555,827 | 2,542,552,916 | 3,338,426 | 3,338,426 | 3,338,427 |
| UPHS | Yes | None | 14,828,230 | 10,917,117,453 | 2,393,946 | 2,393,946 | 5,090,787 |
Intrinsic evaluation
Semantic similarity
The word2vec and fasttext embeddings outperformed the GLoVE embeddings for nearly all corpora and dimension sizes (Figure 1). All of the embeddings trained on clinical corpora outperformed those trained on non-medical text.
Figure 1:
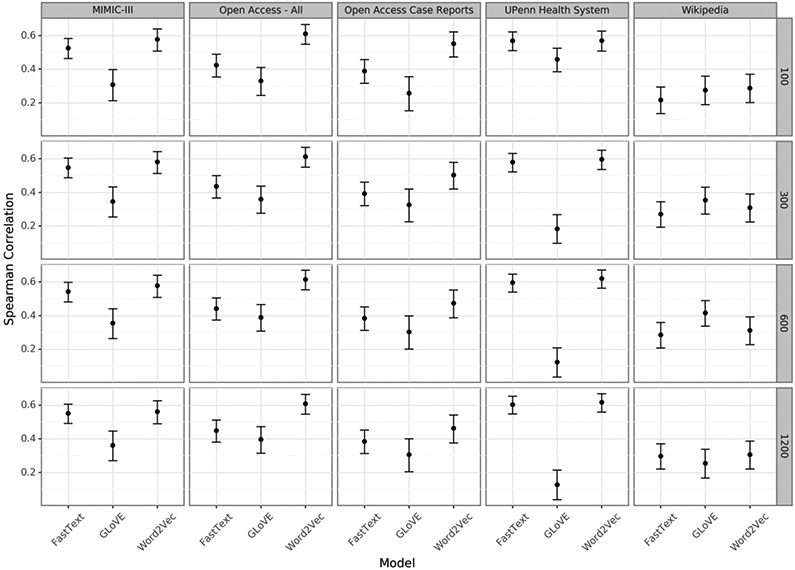
Spearman correlation between the cosine similarity of the words in each pair and the manually annotated similarity.
Linguistic regularity
Word embeddings performed similarly across training models and corpora, but did slightly better with higher vector dimensions (Figure 2, Supplemental Figures 2, 4-5). The GLoVE embeddings performed slightly better for some dimensions with the largest improvements in the smallest corpus. Regularity varied in and between disease types (Supplemental Figures 3, 6-8).
Figure 2:
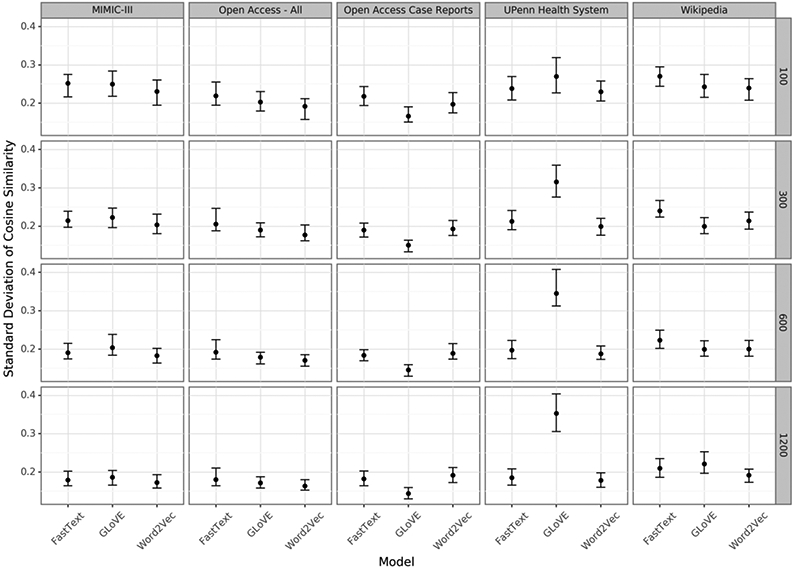
The variance in cosine similarity across vector differences of 100 word pairs related by "is_a_treatment_for". Standard deviation was calculated from the mean cosine similarity of all 100 pairs. Embeddings with lower standard deviation capture a more regular treatment relationship using the same vector difference.
Extrinsic evaluation
Lexicographic coverage
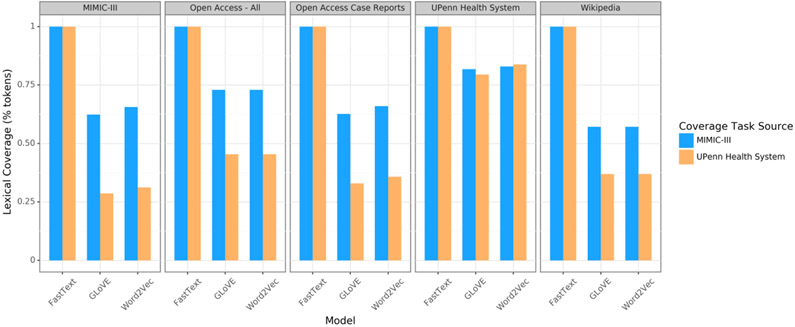
Fasttext embeddings were able to produce vectors for all words while embeddings trained with word2vec and GLoVE not infrequently returned null results for out-of-vocabulary terms (Figure 3). UPHS embeddings had the best coverage across notes from UPHS and from MIMIC-III.
Figure 3:
Fraction of words in a set of clinical encounter notes for which a vector was produced from an embedding set. Intensive care unit encounter notes from the MIMIC-III and University of Pennsylvania Health System datasets were used to measure lexicographic coverage.
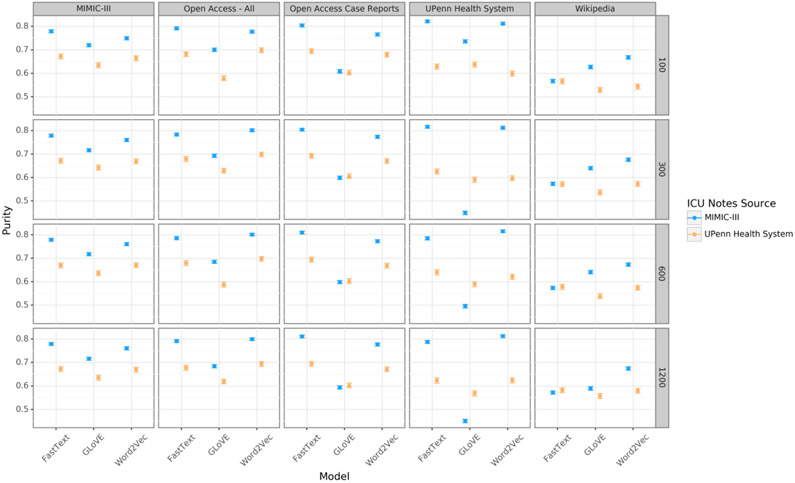
Clustering purity
MIMIC-III discharge summaries were more easily clustered than those from UPHS for nearly all corpora and dimensions (Figure 4). Fasttext embeddings outperformed the other models in almost all cases, and the OA-CR embeddings performed as least as well as the other embeddings in this task. Performance was only minimally affected by the dimension size of the embedding.
Figure 4:
Clustering purity of intensive care unit (ICU) discharge summaries based on a k-means procedure. Discharge summaries from three ICUs in the MIMIC-III dataset and four ICUs in the University of Pennsylvania Health System dataset were used.
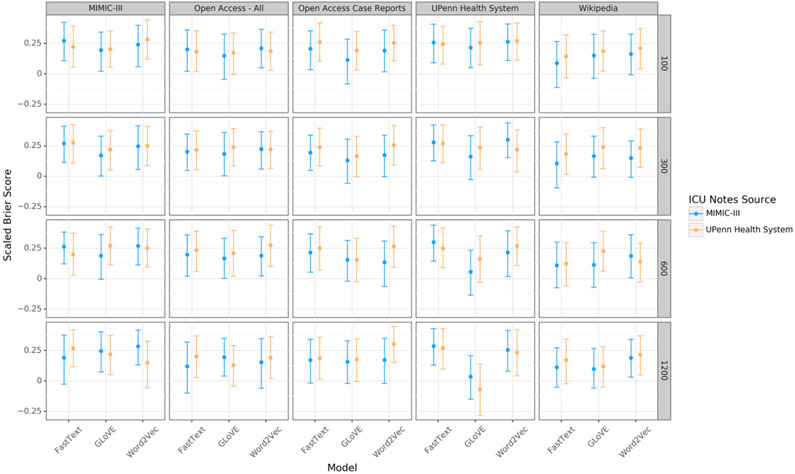
Mortality prediction
Prediction models were fit using 3,336 and 4,122 notes in the MIMIC-III and UPHS datasets respectively. Final performance was reported on the held out test sets that included 834 and 1,030 notes, respectively (Figure 5). Performance ranged from the best model, word2vec OA-CR embeddings with 1,200 dimensional vectors trained on UPHS notes, with a SBS of 0.30 (95% CI 0.15 to 0.45) to the worst, GLoVE UPHS embeddings with 1,200 dimensional vectors trained on UPHS notes, with a SBS of −0.06 (95% CI −0.28 to 0.14). The mortality rate was 12.2% in the MIMIC-III dataset and in the downsampled UPHS dataset.
Figure 5:
Performance of a mortality prediction model using the text of the first physician encounter note for each hospitalization. Performance is reported on a held-out test set the scaled Brier score. Abbreviations: MIMIC-III = Medical Information Mart for Intensive Care III.
De-identification
Wiki embeddings had the highest rate of successful de-identification, while de-identified notes from the UPHS corpus notes trained the best-performing mortality prediction model (Table 3). Relative performance among the embeddings was similar in the i2b2 task (Supplemental Tables 8-9).
Table 3:
De-identification with word embeddings of clinical encounter notes. Intensive care unit notes from the University of Pennsylvania Health System dataset were used for de-identification. De-identified notes were used as input for the mortality prediction task described above and performance is reported as the scaled Brier score. Abbreviations: OA-All = PMC Open Access Subset- All manuscripts, Wiki = Wikipedia-English, UPHS = University of Pennsylvania Health System clinical encounter notes.
| Embedding Set | Notes De-identified (%) | Scaled Brier Score (95% CI) |
|---|---|---|
| OA-All 300d word2vec | 24 (48%) | 0.13 (−0.06, 0.31) |
| UPHS 300d word2vec | 38 (76%) | 0.61 (0.48, 0.73) |
| Wiki 300d word2vec | 39 (78%) | 0.50 (0.37, 0.63) |
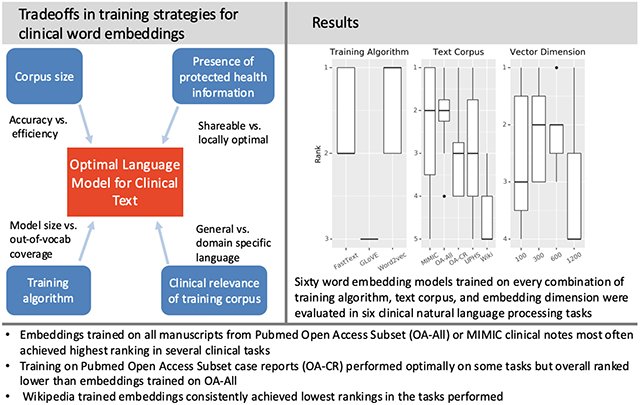
Overall performance rankings
On average, training with word2vec and training on the OA-All and MIMIC corpora produced the best results (Figure 6). Clinical text embeddings outperformed non-clinical text trained embeddings (Supplemental Figure 11), and word-level embeddings more often outperformed sub-word embeddings (Supplemental Figure 12). For individual embedding sets, fasttext trained 100-dimensional vectors trained on the Wikipedia corpus had the highest median performance across tasks but the fasttext trained 1,200-dimensional vectors trained on the OA-All corpus had the most consistently best performance (Supplemental Figures 9-10).
Figure 6:
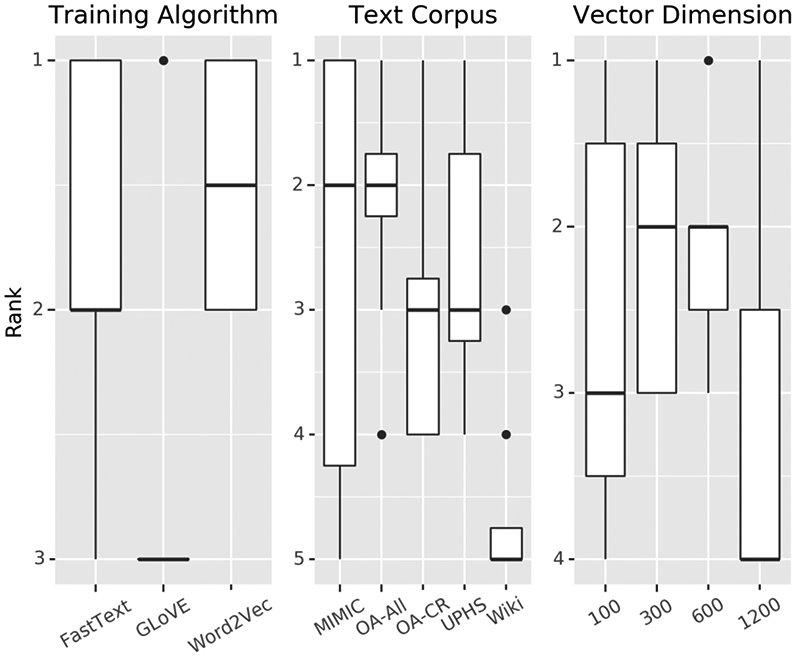
Ranking of task performance across three training variables: training algorithm, text corpus, vector dimension. Performance in each variable was ranked from highest (best performing) to lowest (worst performing).
DISCUSSION
Only minimal differences in performance on intrinsic and extrinsic tasks were identified through comparisons of 60 sets of word embeddings. Consistent with previous studies[1,2] absolute best performance on any given task varied by training algorithm, corpus, and dimension, indicating that no one embedding procedure is optimal. However, there were clear advantages when looking at performance across variable categories (Figure 6). In general, embeddings trained on scientific and medical language corpora outperformed embeddings trained on general language. Wang et al.[2] concluded that general language embeddings were comparable to clinical language embeddings as they observed similar performance in a number of extrinsic NLP tasks, but noted that clinical embeddings did better for semantic tasks. In our analysis, with the exclusion of de-identification, the Wikipedia embeddings were consistently outperformed across the broad array of tasks tested here. This study also confirms Wang’s findings that no single set of embeddings is optimal for all tasks. Additionally, our work extends that of Wang et al. by comparing EHR-based embeddings from two different health systems, neither of which consistently outperformed the other by a meaningful margin. Lastly, by providing results in the form of point estimates with confidence intervals we allow for more nuanced assessment of the relative performance of different training pipelines.
Overall, OA-All and MIMIC embeddings had the best average performance across all tasks, with OA-All embeddings performing more consistently, but the size of the performance differences for most comparisons were small. Word2vec and fasttext embeddings did significantly better than GLoVE embeddings for most tasks. Finally, 300 and 600 dimensional embeddings outperformed smaller and larger dimension embeddings, though the effect was smaller than for corpus and training algorithm. These empirical findings on word embedding performance in clinical NLP tasks have several implications for researchers and data scientists working with clinical text data.
First, publicly accessible, clinical corpora should be used to train embeddings for most clinical tasks. Many previous studies investigating word embeddings for use in clinical NLP have relied on clinical note corpora that are not publicly available.[1,2,13,34,35] Locally trained embeddings create a barrier to the adoption of word embeddings in clinical NLP as such embeddings are not shareable, limiting further evaluation, refinement, and enhancement. Furthermore, the dearth of publicly shareable embeddings, especially ones that have been validated in clinical NLP tasks, functions as a bottleneck for the development of downstream clinical applications that are reproducible. We show that embeddings trained on publicly available clinical text have comparable performance to locally trained embeddings in intrinsic and extrinsic evaluation. Because embeddings trained on the PMC Open Access are completely de-identified through the editorial process, they can be shared. We have made ours available for download (https://github.com/weissman-lab/clinical_embeddings) and we provide a pipeline for updating the embeddings with code available on GitHub. A robust, shareable embedding set allows for overcoming issues of privacy and reproducibility that have hampered the utilization of word embeddings in clinical NLP.
Second, the much smaller OA-CR corpus can be used with minimal performance decrements when computational resources are more limited. Both OA-All and OR-CR embeddings were trained on subsets of the PMC Open Access database, with the OA-CR embeddings being trained exclusively on case reports. The OA-CR embeddings are an order of magnitude smaller than the OA-All embedding (0.7GB vs. 7.8GB for the word2vec 300-dimension embeddings) yet their performance was comparable to OA-All embeddings across most tasks. Both OA-CR and OA-All embeddings are also smaller than existing BioWordVec embeddings for comparable word2vec vectors (300 vs. 200 dimensional vectors respectively) and fasttext embeddings of the same dimensions. The availability of a computationally manageable embedding set will allow for broader incorporation of word embeddings to research projects, especially when access to larger computational resources poses a physical and technical obstacle. It must be noted that the OA-CR embeddings were built for clinical NLP tasks related to understanding and learning from clinical encounter notes. Their effectiveness in other areas of clinical NLP requires further study. As the environmental cost of training large language models increase, using clinically enriched corpora may also represent a more energy efficient solution for clinical tasks.[36]
Third, subword n-grams do not provide dramatic improvements in performance in clinical tasks compared to word-level n-grams. Previously, fasttext embeddings were not found to enhance performance in intrinsic tasks compared to word2vec embeddings.[2,37] However, we hypothesized sub-word embeddings would outperform word-level embeddings in extrinsic tasks where issues of out-of-vocabulary words and misspellings present in real-world EHR data would be manifest. While lexicographic coverage for both MIMIC-III and UPHS ICU notes was 100% for fasttext embeddings and lower for word2vec embeddings, higher coverage did not translate to better performance in ICU class labeling or mortality prediction. This result further complicates issues of clinical text processing and other methods are necessary for dealing with EHR text idiosyncrasies. It is important to note that the extrinsic tasks evaluated here are predictive and it is possible that fasttext based methods for other NLP tasks such as named entity recognition will fare better.
Fourth, embeddings trained on local, PII-containing clinical notes offer minimal performance advantages at the cost of decreased reproducibility. There is good face validity that locally trained embeddings would outperform embeddings trained on public corpora in local tasks by capturing local care patterns and documentation practices. Our UPHS embeddings were trained on one-hundred times the number of encounter notes used in previous studies, and performance in local and non-local NLP tasks was not significantly better, and even worse in some cases. The results for our UPHS embeddings should give pause to researchers believing only locally trained embeddings should be used for their research. However, in de-identification with word embeddings, UPHS embeddings trained on PII-containing text outperformed embeddings trained on non-PII-containing biomedical text. Notably, this is the first study to examine performance of embeddings trained from two different health systems and thus strengthens the case for using open rather than local training corpora. Overall, the Wiki, OA-All, and UPHS corpora contained more biographic syntax necessary for de-identification with word embeddings compared to the smaller, de-identified OA-CR and MIMIC corpora. It is possible there are other local syntactic NLP tasks, such as bias identification in EHR notes, where local training will be produce superior results.
This study should be interpreted in light of several limitations. First, though OA-CR trained embeddings performed very well on average, absolute best performance in a given task varied, indicating that building optimally performing embeddings requires a robust evaluation of embeddings trained on different text corpora. Second, while we evaluated our embeddings in intrinsic and extrinsic tasks, we did not evaluate the embeddings in a clinical named entity recognition task, a popular application of word embeddings,[6,38-40] and it is possible that larger corpora would perform better in a more focused syntactic task. Third, there has been a recent shift in NLP from word embeddings to context embeddings that leverage attention in training[41,42] and it remains to be seen if context embeddings built from published case reports can provide the same performance, though contextual embeddings have only showed a small performance increase compared to word embeddings in clinical NLP tasks including entity recognition[43], semantic similarity[37], and disease prediction.[44] While context-sensitive embeddings such as BERT offer promising results for NLP broadly, training from scratch requires a substantially higher computational cost[40] and further investigations of pre-trained clinical models[37,44] are warranted. Additionally, the environmental impact of training large models warrants careful consideration of long-term sustainability and benefits.[36] Finally, the inferential and explanatory relevance of offset vectors may be confounded by corpus-level noise and other irregularities.[45,46] Therefore, further work is needed to test the clinical relevance of analogy completion in downstream reasoning tasks and validate the appropriateness of such tasks for evaluating clinical corpora.
CONCLUSION
There are a number of recognized limitations to the use of word embeddings for clinical text representation including methods of validation and issues relating to sharing embedding sets such as interoperability and privacy.[13] We sought to address these issues by building and evaluating embeddings trained on publicly available clinical case reports. These embeddings perform comparably to embeddings trained from general and clinical text corpora in a variety of intrinsic and extrinsic tasks, even at a fraction of the training corpus size. We make these embeddings available for download to alleviate researchers of having to construct their own embedding as well as for benchmarking new embedding sets and embedding methods. Training local embeddings requires access to clinical text, computational resources, and methods of validation. Such a high upfront cost has prevented the utilization of word embeddings in clinical research projects. We hope to facilitate their use by providing a downloadable set of word embeddings that can be reproduced, updated, and has been experimentally validated.
Supplementary Material
Highlights.
Published case report text approximates clinical notes for language model training
Training with case reports performs comparably to training on larger corpora
Subword training does not increase performance in clinical text prediction
Training on text containing protected health information offers minimal benefit
Funding
This work was supported by the National Institutes of Health [NIH/NHLBI K23HL141639] and the Penn Center for Precision Medicine, Philadelphia, PA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
Publicly shareable word embedding models are available at https://github.com/weissman-lab/clinical_embeddings. Datasets derived from sources in the public domain are: Pubmed Open Access, MIMIC-III, and Wikipedia. UPHS data cannot be shared for privacy reasons because they contain protected health information. UPHS data was provided under IRB approval from the University of Pennsylvania.
REFERENCES
- 1.Pakhomov SV, Finley G, McEwan R et al. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics 2016;btw529. doi: 10.1093/bioinformatics/btw529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Liu S, Afzal N et al. A comparison of word embeddings for the biomedical natural language processing. Journal of Biomedical Informatics 2018;87:12–20. doi: 10.1016/j.jbi.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts K Assessing the corpus size vs. similarity trade-off for word embeddings in clinical NLP. In: Proceedings of the clinical natural language processing workshop (ClinicalNLP). Osaka, Japan:: The COLING 2016 Organizing Committee; 2016. 54–63.https://www.aclweb.org/anthology/W16-4208.pdf [Google Scholar]
- 4.Abdalla M, Abdalla M, Hirst G et al. Exploring the privacy-preserving properties of word embeddings: Algorithmic validation study. Journal of Medical Internet Research Published Online First: 2020. doi: 10.2196/18055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beam AL, Kompa B, Schmaltz A et al. Clinical concept embeddings learned from massive sources of multimodal medical data. In: Biocomputing 2020.295–306. doi: 10.1142/9789811215636_0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu B, Crichton G, Korhonen A et al. How to train good word embeddings for biomedical NLP. In: Proceedings of the 15th workshop on biomedical natural language processing. Berlin, Germany:: Association for Computational Linguistics; 2016. 166–74. doi: 10.18653/v1/W16-2922 [DOI] [Google Scholar]
- 7.TH M, Sahu S, Anand A. Evaluating distributed word representations for capturing semantics of biomedical concepts. In: Proceedings of BioNLP 15. Beijing, China:: Association for Computational Linguistics; 2015. 158–63. doi: 10.18653/v1/W15-3820 [DOI] [Google Scholar]
- 8.Huang J, Xu K, Vydiswaran VGV. Analyzing multiple medical corpora using word embedding. 2016;527–33. doi: 10.1109/ICHI.2016.94 [DOI] [Google Scholar]
- 9.Chen Z, He Z, Liu X et al. Evaluating semantic relations in neural word embeddings with biomedical and general domain knowledge bases. BMC Medical Informatics and Decision Making Published Online First: 2018. doi: 10.1186/s12911-018-0630-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Major V, Surkis A, Aphinyanaphongs Y. Utility of General and Specific Word Embeddings for Classifying Translational Stages of Research. AMIA Annual Symposium Proceedings 2018;2018:1405–14. [PMC free article] [PubMed] [Google Scholar]
- 11.Boag W, Doss D, Naumann T et al. What’s in a Note? Unpacking Predictive Value in Clinical Note Representations. AMIA Summits on Translational Science Proceedings 2018;2017:26–34. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, He Z, Liu X et al. Evaluating semantic relations in neural word embeddings with biomedical and general domain knowledge bases. BMC Medical Informatics and Decision Making 2018;18. doi: 10.1186/s12911-018-0630-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khattak FK, Jeblee S, Pou-Prom C et al. A survey of word embeddings for clinical text. Journal of Biomedical Informatics: X 2019;4:100057. doi: 10.1016/j.yjbinx.2019.100057 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Chen Q, Yang Z et al. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Scientific Data 2019;6:1–9. doi: 10.1038/s41597-019-0055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdalla M, Abdalla M, Rudzicz F et al. Using word embeddings to improve the privacy of clinical notes. Journal of the American Medical Informatics Association 2020;27:901–7. doi: 10.1093/jamia/ocaa038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bojanowski P, Grave E, Joulin A et al. Enriching word vectors with subword information. Published Online First: 2017.https://arxiv.org/abs/1607.04606 [Google Scholar]
- 17.Mikolov T, Chen K, Corrado G et al. Efficient estimation of word representations in vector space. Published Online First: January 2013.https://arxiv.org/pdf/1301.3781.pdf [Google Scholar]
- 18.Keselman A, Smith CA. A classification of errors in lay comprehension of medical documents. Journal of Biomedical Informatics Published Online First: 2012. doi: 10.1016/j.jbi.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuyo N Language detection library for java. 2010.http://code.google.com/p/language-detection/ [Google Scholar]
- 20.Johnson AE, Pollard TJ, Shen L et al. MIMIC-III, a freely accessible critical care database. Scientific Data 2016;3:160035. doi: 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberger AL, Amaral LAN, Glass L et al. PhysioBank, physiotoolkit, and physionet. Circulation 2000;101:e215–20. doi: 10.1161/01.CIR.101.23.e215 [DOI] [PubMed] [Google Scholar]
- 22.Rehuřek R, Sojka P. Software Framework for Topic Modelling with Large Corpora. In: Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks. Valletta, Malta:: ELRA; 2010.45–50. [Google Scholar]
- 23.Bird S, Klein E, Loper E. Natural language processing with python. 1st ed. O’Reilly Media, Inc. 2009. [Google Scholar]
- 24.Medicine (U.S.) NL of. UMLS knowledge sources: Metathesaurus, semantic network, [and] specialist lexicon. U.S. Department of Health; Human Services, National Institutes of Health, National Library of Medicine; 2003. https://books.google.com/books?id=xTtrAAAAMAAJ [Google Scholar]
- 25.Singh V Replace or retrieve keywords in documents at scale. Published Online First: 2017.https://arxiv.org/abs/1711.00046 [Google Scholar]
- 26.Honnibal M, Johnson M. An improved non-monotonic transition system for dependency parsing. In: Proceedings of the 2015 conference on empirical methods in natural language processing. Lisbon, Portugal:: Association for Computational Linguistics; 2015. 1373–8.https://aclweb.org/anthology/D/D15/D15-1162 [Google Scholar]
- 27.Pennington J, Socher R, Manning C. Glove: Global Vectors for Word Representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Doha, Qatar:: Association for Computational Linguistics; 2014. 1532–43. doi: 10.3115/v1/D14-1162 [DOI] [Google Scholar]
- 28.Pakhomov S, McInnes B, Adam T et al. Semantic Similarity and Relatedness between Clinical Terms: An Experimental Study. AMIA Annual Symposium Proceedings 2010;2010:572–6. [PMC free article] [PubMed] [Google Scholar]
- 29.Mikolov T, Yih W-t, Zweig G. Linguistic regularities in continuous space word representations. In: Proceedings of the 2013 conference of the north american chapter of the association for computational linguistics: Human language technologies. 2013. 746–51. [Google Scholar]
- 30.Zhang Z, Hong Y. Development of a novel score for the prediction of hospital mortality in patients with severe sepsis: The use of electronic healthcare records with lasso regression. Oncotarget 2017;8:49637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y. Convolutional neural networks for sentence classification. Published Online First: 2014.https://arxiv.org/abs/1408.5882 [Google Scholar]
- 32.Steyerberg EW, Vickers AJ, Cook NR et al. Assessing the performance of prediction models: A framework for traditional and novel measures. 2010. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubbs A, Uzuner Ö. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/uthealth corpus. Journal of Biomedical Informatics 2015;58:S20–9. doi: 10.1016/j.jbi.2015.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen P, Tran T, Wickramasinghe N et al. Deepr: A convolutional net for medical records. 2016.https://arxiv.org/abs/1607.07519 [DOI] [PubMed] [Google Scholar]
- 35.Craig E, Arias C, Gillman D. Predicting readmission risk from doctors’ notes.2017.https://arxiv.org/abs/1711.10663 [Google Scholar]
- 36.Strubell E, Ganesh A, McCallum A. Energy and policy considerations for deep learning in nlp. 2019.https://arxiv.org/abs/1906.02243 [Google Scholar]
- 37.Huang K, Altosaar J, Ranganath R. ClinicalBERT: Modeling clinical notes and predicting hospital readmission. 2020.https://arxiv.org/abs/1904.05342 [Google Scholar]
- 38.Lee J, Yoon W, Kim S et al. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics Published Online First: September 2019. doi: 10.1093/bioinformatics/btz682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltagy I, Lo K, Cohan A. SciBERT: A pretrained language model for scientific text. 2019.https://arxiv.org/abs/1903.10676 [Google Scholar]
- 40.Alsentzer E, Murphy J, Boag W et al. Publicly available clinical BERT embeddings. In: Proceedings of the 2nd clinical natural language processing workshop. Minneapolis, Minnesota, USA:: Association for Computational Linguistics; 2019. 72–8. doi: 10.18653/v1/W19-1909 [DOI] [Google Scholar]
- 41.Vaswani A, Shazeer N, Parmar N et al. Attention is all you need. 2017.https://arxiv.org/abs/1706.03762 [Google Scholar]
- 42.Devlin J, Chang M-W, Lee K et al. BERT: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 conference of the north American chapter of the association for computational linguistics: Human language technologies, volume 1 (long and short papers). Minneapolis, Minnesota:: Association for Computational Linguistics; 2019. 4171–86. doi: 10.18653/v1/N19-1423 [DOI] [Google Scholar]
- 43.Si Y, Wang J, Xu H et al. Enhancing clinical concept extraction with contextual embeddings. Journal of the American Medical Informatics Association 2019;26:1297–304. doi: 10.1093/jamia/ocz096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmy L, Xiang Y, Xie Z et al. Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. npj Digital Medicine 2021;4:86. doi: 10.1038/s41746-021-00455-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers A, Drozd A, Li B. The (too many) problems of analogical reasoning with word vectors. In: Proceedings of the 6th joint conference on lexical and computational semantics (*SEM 2017). Vancouver, Canada:: Association for Computational Linguistics; 2017. 135–48. doi: 10.18653/v1/S17-1017 [DOI] [Google Scholar]
- 46.Linzen T Issues in evaluating semantic spaces using word analogies. In: Proceedings of the 1st workshop on evaluating vector-space representations for NLP. Berlin, Germany:: Association for Computational Linguistics; 2016. 13–8. doi: 10.18653/v1/W16-2503 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly shareable word embedding models are available at https://github.com/weissman-lab/clinical_embeddings. Datasets derived from sources in the public domain are: Pubmed Open Access, MIMIC-III, and Wikipedia. UPHS data cannot be shared for privacy reasons because they contain protected health information. UPHS data was provided under IRB approval from the University of Pennsylvania.