Figure 1. The DNA methylation-demethylation cycle.
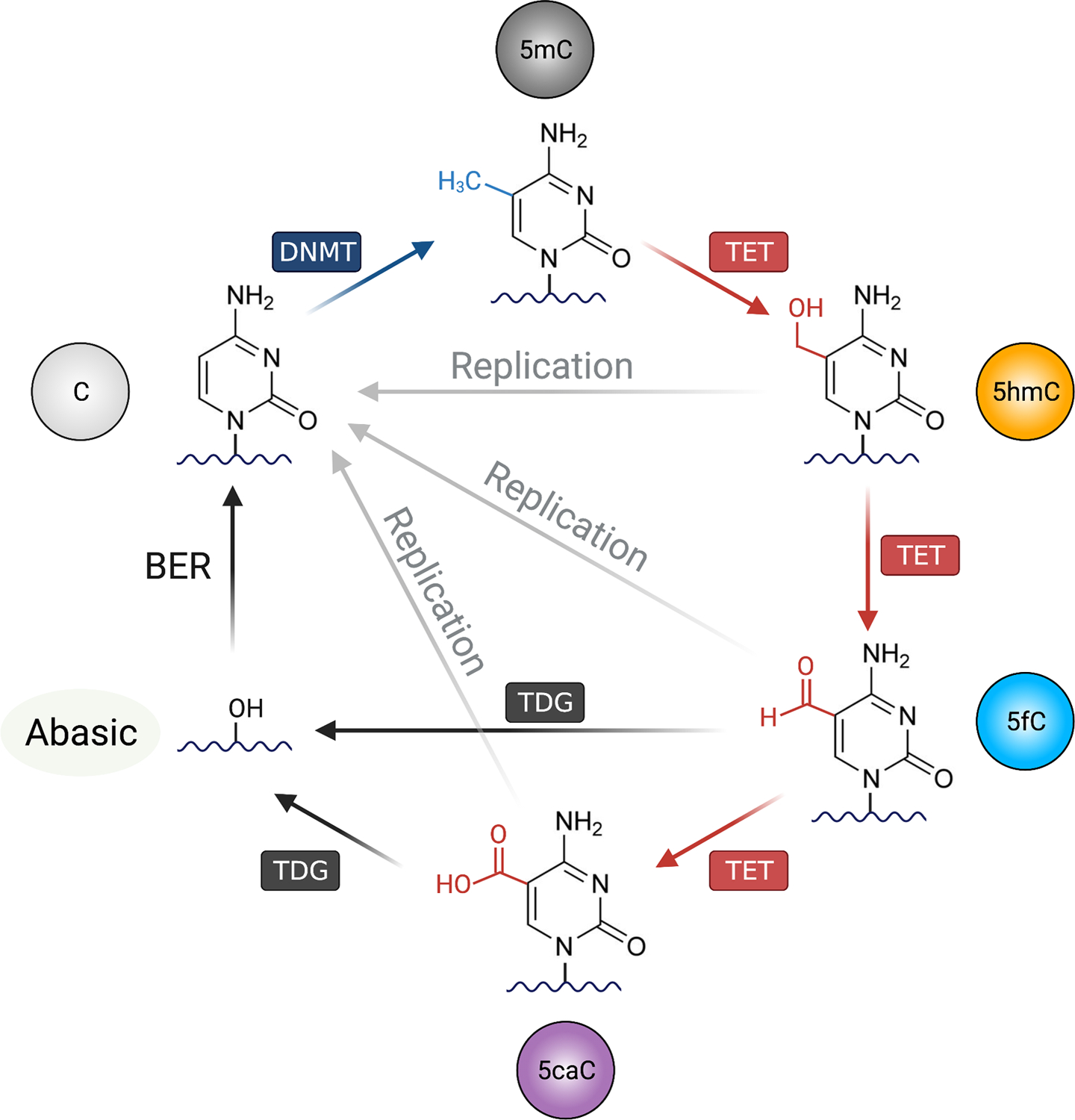
Unmodified cytosine (C) on the DNA can be methylated by the DNA methyltransferases (DNMTs), forming 5-methylcytosine (5mC). For DNA demethylation, TET (Ten-Eleven Translocation) oxidizes 5mC into oxidized cytosines including 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). There are mainly two mechanisms for TET-mediated DNA demethylation: replication-independent (active) and replication-dependent (passive) pathways. In active DNA demethylation, 5fC and 5caC are recognized and excised by thymine DNA glycosylase (TDG), creating an abasic site that is replaced with unmethylated cytosine by base excision repair (BER) pathway. In passive demethylation, while the DNMT1/UHRF1 complex recognize hemi-methylated DNA CpG on replicating DNA, the complex is not able to recognize and methylate the hemi-methylated CpG containing oxidized cytosines (5fC, 5fC and 5caC), thus inhibiting the methylation of the CpG motif on the newly replicated DNA. Figure was created with BioRender.
