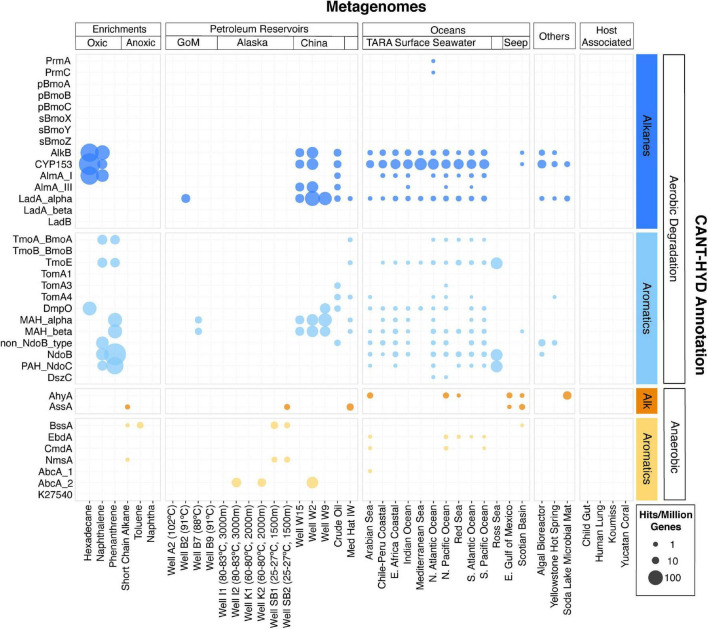
FIGURE 6.
Metagenomes from diverse environments analyzed with the CANT-HYD HMMs. CANT-HYD HMM hits (scores ≥ noise cutoff) (y-axis) to each metagenome (x-axis) normalized to per million protein coding genes. The size of the bubble depicts proportion of hydrocarbon degrading genes to total genic content of the metagenome. The metagenomes (x-axis) are grouped under Enrichments (hydrocarbon-degrading enrichment cultures), Petroleum Reservoirs (produced well water from petroleum reservoirs from Alaska, Gulf of Mexico, Jiangsu and Qinghai, China, and Medicine Hat, Alberta), Oceans (cold seeps, TARA surface seawater), Host-Assoc (host-associated microbiomes), Other: (other environments). The CAN-HYD HMMs are grouped by hydrocarbon substrate (alkane and aromatic) and respiration (aerobic and anaerobic). See Supplementary Table 4 for details regarding these metagenomes.