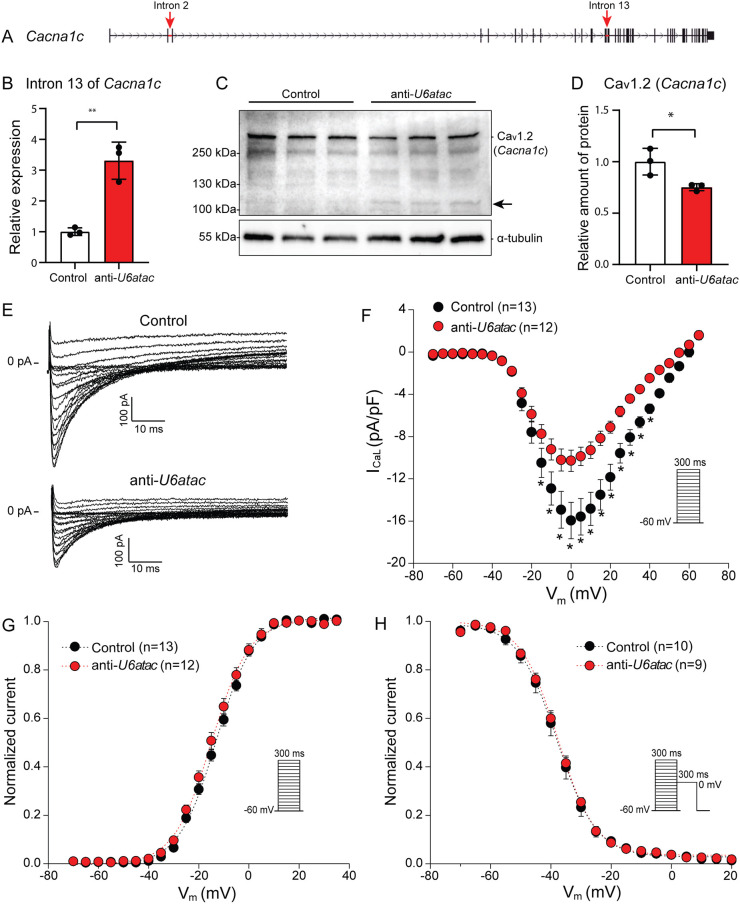
Fig. 5.
Minor intron retention of Cacna1c hampers translation into Cav1.2 and reduces ICaL density. (A) Gene structure of Cacna1c with minor introns indicated with red arrows. (B) Minor intron retention in Cacna1c after U6atac knockdown measured by qRT-PCR. (C) Cav1.2 (Cacna1c) detection by western blot and (D) its quantification. Black arrow indicates a potential truncated isoform of Cav1.2. Data in B,D are presented as mean±s.d., 3 wells per condition. *P<0.05, **P<0.01 (unpaired two-tailed Student's t-test). (E) Representative ICaL traces recorded from NRVMs co-transfected with control or anti-U6atac gapmeR. (F–H) Average ICaL–voltage relationships (F), ICaL-voltage dependence of activation (G) and inactivation (H). Insets show voltage protocols. Patch-clamp data are presented as mean±s.e.m.; n indicates number of cardiomyocytes. *P<0.05 (two-way repeated measures ANOVA followed by Holm–Sidak test for post hoc analyses).