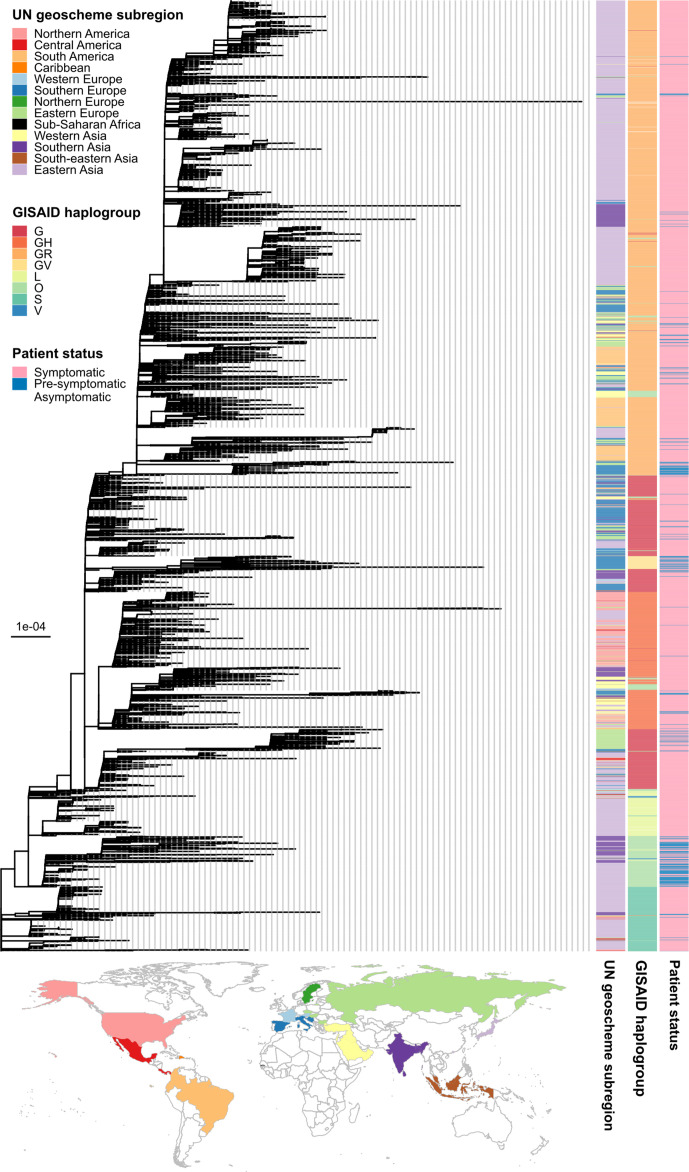
Fig. 1.
SARS-CoV-2 phylogeny. The tree was estimated under the maximum-likelihood framework implemented in IQ-TREE2 [27] based on a manually curated alignment of 3021 full-length SARS-CoV-2 genomes. Potential recombination within the alignment was checked by using the Phi test implemented in SplitsTree4 [26], but no evidence was found (P=0.91). The best-fit nucleotide substitution model was determined to be GTR+F+R5 (the general time reversible model+empirical base frequencies+the 5-discrete-rate-category FreeRate model) by ModelFinder [28] under the Bayesian information criterion and was used for tree reconstruction. We compared our tree with the global tree obtained from GISAID, and determined the terminal branch leading to sample EPI_ISL_407976 as a suitable location for root placement. Bar, substitutions per site. The tree file in Newick format with bootstrap clade-support values, computed based on 1000 bootstrap trees, can be found in Data S1. The three columns on the right indicate the United Nations (UN) geoscheme subregion, the GISAID haplogroup assignment and the patient status of the sequences, respectively (see keys). The World map below the tree shows the countries from which the sequences were sampled, colored according to the UN geoscheme subregions.