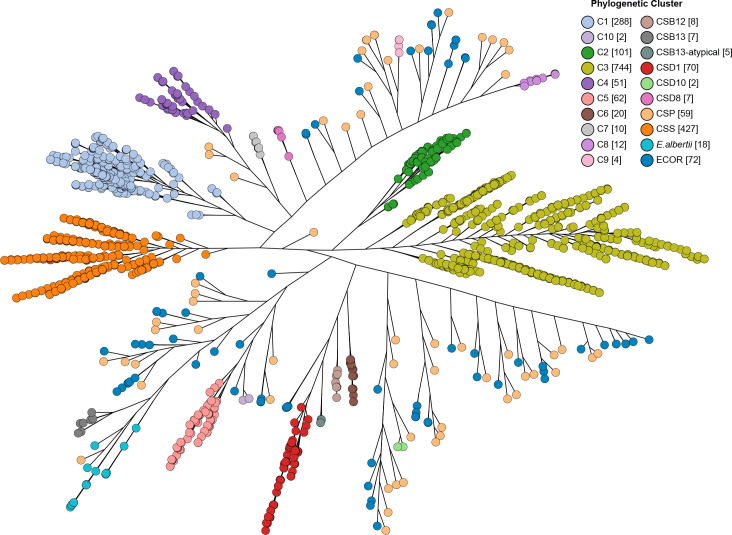
Fig. 1.
Shigella and EIEC cluster identification phylogenetic tree. Representative isolates from the identification dataset were used to construct the phylogenetic tree using Quicktree v1.3 [65] to identify Shigella and EIEC (enteroinvasive E. coli ) clusters and visualized using Grapetree. The dendrogram shows the phylogenetic relationships of 1879 Shigella and EIEC isolates represented in the identification dataset. Branch lengths are on a log scale for clarity. Bar, 0.2 substitutions per site. Shigella and EIEC clusters are coloured. Numbers in square brackets after the cluster name are the number of isolates for each identified cluster. CSP indicates sporadic EIEC lineages. ECOR is the Escherichia coli reference collection. E. albertii is Escherichia albertii which was included to show the location of ‘typical’ S. boydii serotype 13 strains. CSS, CSB12, CSB13, CSD1, CSD8 and CSD10 are the clusters of S. sonnei , S. boydii serotype 12, S. boydii serotype 13, S. dysenteriae serotype 1, S. dysenteriae serotype 8 and S. dysenteriae serotype 10 respectively.