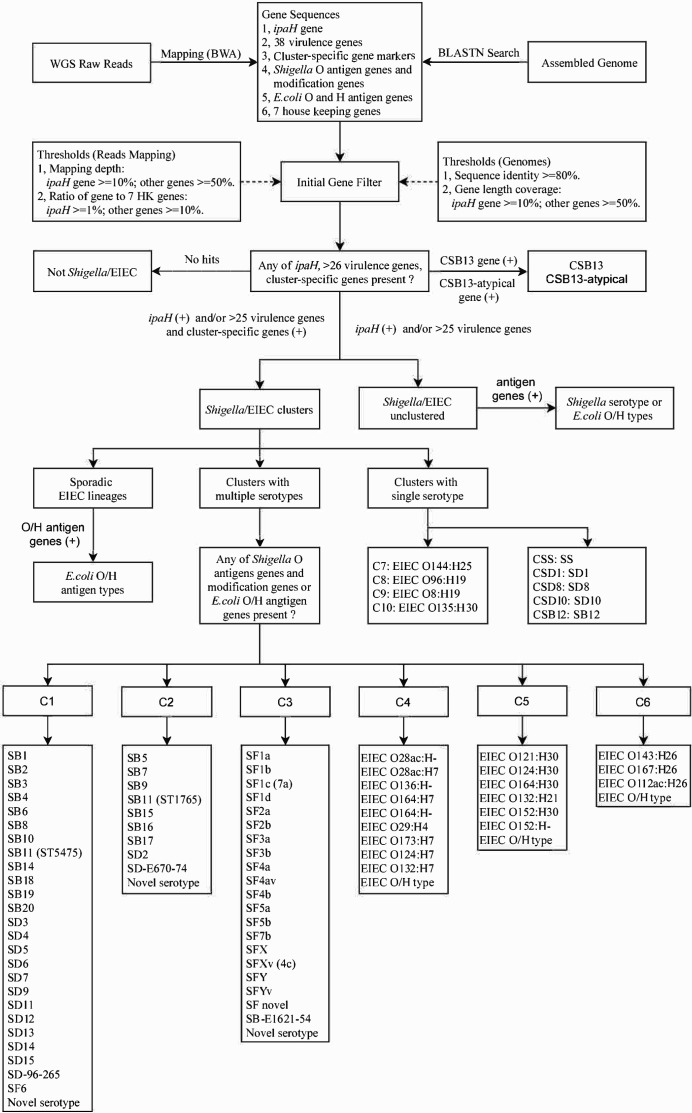
Fig. 2.
In silico serotyping pipeline workflow. Schematic of in silico serotyping of Shigella and EIEC (enteroinvasive E. coli ) by cluster-specific genes combined with the ipaH gene, O antigen and modification genes and H antigen genes, implemented in ShigEiFinder. Both assembled genomes and raw reads are accepted as data input. The dotted arrows show the cutoff value applied for initial gene filtering. WGS, whole-genome sequencing; HK, housekeeping; SS, SF, SB and SD, S. sonnei , S. flexneri , S. boydii and S. dysenteriae respectively. The abbreviated ‘species’ name plus the serotype number is the designation of a Shigella serotype (e.g. S. dysenteriae serotype 1 is abbreviated as SD1). For SB11, there were two sequence types (STs) with ST5475 and ST1765 located within clusters C1 and C2 respectively.