Abstract
The emergence of the highly transmissible B.1.1.529 Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is concerning for antibody countermeasure efficacy because of the number of mutations in the spike protein. In this study, we tested a panel of anti-receptor-binding domain monoclonal antibodies (mAbs) corresponding to those in clinical use by Vir Biotechnology (S309, the parent mAb of VIR-7831 (sotrovimab)), AstraZeneca (COV2-2196 and COV2-2130, the parent mAbs of AZD8895 and AZD1061), Regeneron (REGN10933 and REGN10987), Eli Lilly (LY-CoV555 and LY-CoV016) and Celltrion (CT-P59) for their ability to neutralize an infectious B.1.1.529 Omicron isolate. Several mAbs (LY-CoV555, LY-CoV016, REGN10933, REGN10987 and CT-P59) completely lost neutralizing activity against B.1.1.529 virus in both Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells, whereas others were reduced (COV2-2196 and COV2-2130 combination, ~12-fold decrease) or minimally affected (S309). Our results suggest that several, but not all, of the antibodies in clinical use might lose efficacy against the B.1.1.529 Omicron variant.
Subject terms: SARS-CoV-2, Viral immune evasion
New in vitro data suggest that the new SARS-CoV-2 Omicron variant is likely to escape neutralization by most therapeutic antibodies currently available.
Main
Since December 2019, the global Coronavirus Disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has resulted in 298 million infections and 5.4 million deaths. The expansion of the COVID-19 pandemic and its accompanying morbidity, mortality and destabilizing socioeconomic effects have made the development and distribution of SARS-CoV-2 therapeutics and vaccines an urgent global health priority1. Although the rapid deployment of countermeasures, including mAbs and multiple highly effective vaccines, has provided hope for curtailing disease and ending the pandemic, this has been jeopardized by the emergence of more transmissible variants with mutations in the spike protein that also could evade protective immune responses.
Indeed, over the past year, several variant strains have emerged, including B.1.1.7 (Alpha), B.1.351 (Beta), B.1.1.28 (also called P.1, Gamma) and B.1.617.2 (Delta), among others, each having varying numbers of substitutions in the N-terminal domain (NTD) and the receptor-binding domain (RBD) of the SARS-CoV-2 spike. Cell-based assays with pseudoviruses or authentic SARS-CoV-2 strains suggest that neutralization by many Emergency Use Authorization (EUA) mAbs might be diminished against some of these variants, especially those containing mutations at positions L452, K477 and E484 (refs. 2–6). Notwithstanding this, in vivo studies in animals showed that, when most EUA mAbs were used in combination, they retained efficacy against different variants7. The recent emergence of B.1.1.529, the Omicron variant8,9, which has a larger number of mutations (>30 substitutions, deletions or insertions) in the spike protein, has raised concerns that this variant will escape from protection conferred by vaccines and therapeutic mAbs.
Results
We obtained an infectious clinical isolate of B.1.1.529 from a symptomatic individual in the United States (hCoV-19/USA/WI-WSLH-221686/2021). We propagated the virus once in Vero cells expressing human transmembrane protease serine 2 (TMPRSS2) to prevent the emergence of adventitious mutations at or near the furin cleavage site in the spike protein10. Our B.1.1.529 isolate encodes the following mutations in the spike protein (A67V, Δ69−70, T95I, G142D, Δ143-145, Δ211, L212I, insertion 214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K and L981F; Fig. 1a,b and GISAID: EPI_ISL_7263803), which is similar to strains identified in Africa11. Our isolate, however, lacks an R346K mutation, which is present in a minority (~8%) of reported strains.
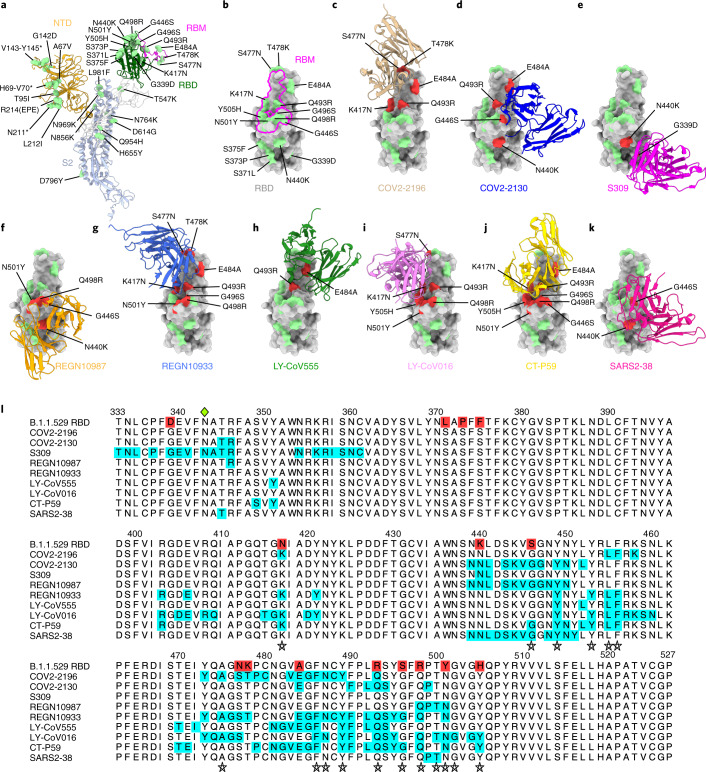
Fig. 1. Neutralizing mAb epitopes on B.1.1.529.
a, b, SARS-CoV-2 spike trimer (PDB: 7C2L and PDB: 6W41). One spike protomer is highlighted, showing the NTD in orange, RBD in green, RBM in magenta and S2 portion of the molecule in blue (a). Close-up view of the RBD with the RBM outlined in magenta (b). Amino acids that are changed in B.1.1.529 compared to WA1/2020 are indicated in light green (a, b), with the exception of N679K and P681H, which were not modeled in the structures used. c–k, SARS-CoV-2 RBD bound by EUA mAbs COV2-2196 (c, PDB: 7L7D); COV2-2130 (d, PDB: 7L7E); S309 (e, PDB: 6WPS); REGN10987 (f, PDB: 6XDG); REGN10933 (g, PDB: 6XDG); LY-CoV555 (h, PDB: 7KMG); LY-CoV016 (i, PDB: 7C01); CT-P59 (j PDB: 7CM4); and SARS2-38 (k, PDB: 7MKM). Residues mutated in the B.1.1.529 RBD and contained in these mAbsʼ respective epitopes are shaded red, whereas those outside the epitope are shaded green. l, Multiple sequence alignment showing the epitope footprints of each EUA mAb on the SARS-CoV-2 RBD highlighted in cyan. B.1.1.529 RBD is shown in the top row, with sequence changes relative to the wild-type RBD highlighted red. A green diamond indicates the location of the N-linked glycan at residue 343. Stars below the alignment indicate hACE2 contact residues on the SARS-CoV-2 RBD40.
Given the number of substitutions in the B.1.1.529 spike protein, including eight amino acid changes (K417N, G446S, S477N, Q493R, G496S, Q498R, N501Y and Y505H) in the ACE2 receptor-binding motif (RBM), we first evaluated possible effects on the structurally defined binding epitopes12,13 of mAbs corresponding to those with EUA approval or in advanced clinical development (S309 (parent of VIR-7831 (sotrovimab)), RBD group III)14,15; COV2-2196 (RBD group I) and COV2-2130 (RBD group III) (parent mAbs of AZD8895 and AZD1061, respectively)16; REGN10933 (RBD group I) and REGN10987 (RBD group III)17; LY-CoV555 (RBD group I) and LY-CoV016 (RBD group I)18,19; and CT-P59 (Celltrion, RBD group I)20, along with an additional broadly neutralizing mAb (SARS2-38 (RBD group II)) that we recently described21. We mapped the B.1.1.529 spike mutations onto the antibody-bound SARS-CoV-2 spike or RBD structures published in the RCSB Protein Data Bank (PDB) (Fig. 1c–k). Although every antibody analyzed had structurally defined recognition sites that were altered in the B.1.1.529 spike, the differences varied among mAbs, with some showing larger numbers of changed residues (Fig. 1l: COV2-2196, n = 5; COV2-2130, n = 4; S309, n = 2; REGN10987, n = 4; REGN10933, n = 8; Ly-CoV555, n = 2; Ly-CoV016, n = 6; CT-P59, n = 8; and SARS2-38, n = 2).
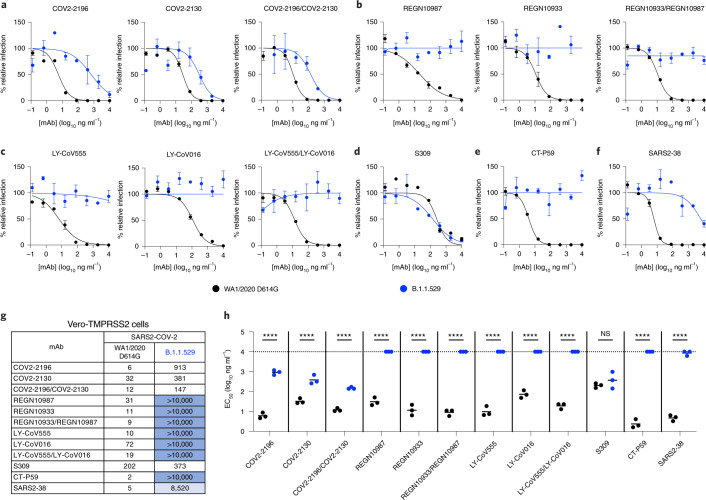
To address the functional significance of the spike sequence variation in B.1.1.529 for antibody neutralization, we used a high-throughput focus reduction neutralization test (FRNT)22 with WA1/2020 D614G and B.1.1.529 in Vero-TMPRSS2 cells (Fig. 2). We tested individual mAbs and combinations of mAbs that target the RBD in Vero-TMPRSS2 cells, including S309 (Vir Biotechnology); COV2-2130/COV2-2196 (parent mAbs of AZD1061 and AZD8895, provided by Vanderbilt University Medical Center); REGN10933/REGN10987 (synthesized based on casirivimab and imdevimab sequences from Regeneron); LY-CoV555/LY-CoV016 (synthesized based on bamlanivimab and etesevimab sequences from Eli Lilly); CT-P59 (synthesized based on regdanvimab sequences from Celltrion); and SARS2-38. As expected, all individual mAbs or combinations of mAbs tested neutralized the WA1/2020 D614G isolate, with half-maximal inhibitory concentration (EC50) values similar to published data6,20,23. However, when tested alone, REGN10933, REGN10987, LY-CoV555, LV-CoV016, CT-P59 and SARS2-38 completely lost neutralizing activity against B.1.1.529, with little inhibitory capacity even at the highest (10,000 ng ml−1) concentration tested. COV2-2130 and COV2-2196 showed an intermediate ~12-fold and 150-fold (P < 0.0001) loss in inhibitory activity, respectively, against the B.1.1.529 strain. In comparison, S309 showed a less than two-fold (P > 0.5) reduction in neutralizing activity against B.1.1.529 (Fig. 2a–h). Analysis of mAb combinations currently in clinical use showed that REGN10933/REGN10987 and LY-CoV555/LV-CoV016 lost all neutralizing activity against B.1.1.529, whereas COV2-2130/COV2-2196 showed a ~12-fold (P < 0.0001) reduction in inhibitory activity from an EC50 of 12 ng ml−1 to 147 ng ml−1.
Fig. 2. Neutralization of SARS-CoV-2 B.1.1.529 Omicron strain by mAbs in Vero-TMPRSS2 cells.
a–f, Neutralization curves in Vero-TMPRSS2 cells comparing the sensitivity of SARS-CoV-2 strains with the indicated mAbs (COV2-2196, COV2-2130, REGN10933, REGN10987, LY-CoV555, LY-CoV016, S309, CT-P59 and SARS2-38) with WA1/2020 D614G and B.1.1.529. Also shown are the neutralization curves for antibody cocktails (COV2-2196/COV2-2130, REGN10933/REGN10987 or LY-CoV555/LY-CoV016). For data with mAb combinations, the x axis represents the total concentration of mAb used. One representative experiment of three performed in technical duplicate is shown. Error bars indicate the range of technical replicates. Data (% relative infection) are normalized to a no-mAb control. g, Summary of EC50 values (ng ml−1) of neutralization of SARS-CoV-2 viruses (WA1/2020 D614G and B.1.1.529) performed in Vero-TMPRSS2 cells. Data are the geometric mean of three experiments. Blue shading: light, EC50 > 5,000 ng ml−1; dark, EC50 > 10,000 ng ml−1. h, Comparison of EC50 values by mAbs against WA1/2020 D614G and B.1.1.529 (three experiments; NS, not significant; ****P < 0.0001; two-way ANOVA with Sidak’s post test). Each symbol represents neutralization data from an individual experiment. Bars indicate mean values. The dotted line indicates the upper limit of dosing of the assay.
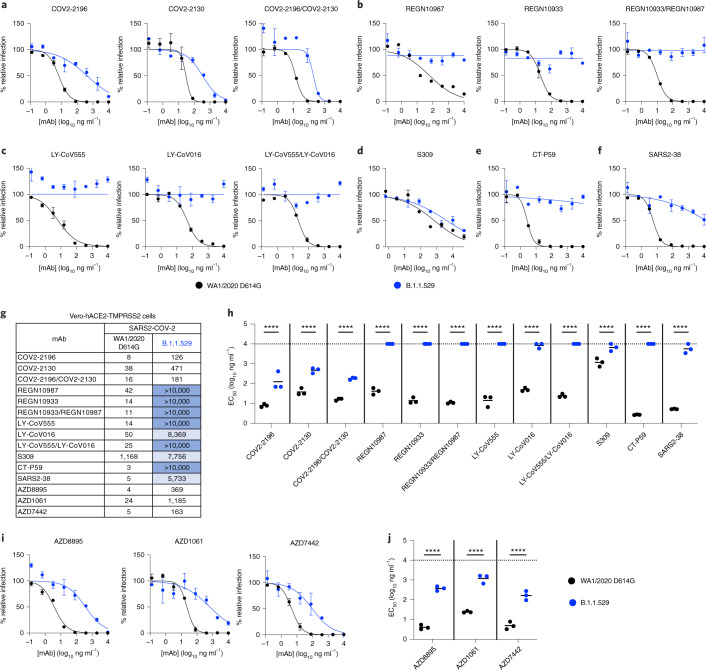
We repeated experiments in Vero-hACE2-TMPRSS2 cells to account for effects of hACE2 expression, which can affect neutralization by some anti-SARS-CoV-2 mAbs21,24. Moreover, modeling studies suggest that the mutations in the B.1.1.529 spike might enhance interactions with hACE2 (ref. 25). All individual mAbs or combinations of mAbs tested neutralized the WA1/2020 D614G isolate as expected. However, REGN10933, REGN10987, LY-CoV555, LV-CoV016, SARS2-38 and CT-P59 completely lost neutralizing activity against B.1.1.529, and the combinations of REGN10933/REGN10987 or LY-CoV555/LV-CoV016 also lacked inhibitory capacity (Fig. 3a–h). In comparison, COV2-2130 and COV2-2196 showed reduced activity (~12-fold and 16-fold, respectively, P < 0.0001) against B.1.1.529, as did the combination of COV2-2130/COV2-2196 mAbs (~11-fold, P < 0.0001). The S309 mAb exhibited less potent neutralizing activity in Vero-hACE2-TMPRSS2 cells against WA1/2020 D614G virus with a flatter dose–response curve (Fig. 3d), as seen previously6,26, and showed a moderate (~six-fold, P < 0.0001) reduction in neutralizing activity against B.1.1.529. Thus, although the trends in mAb neutralization of B.1.1.529 generally were similar to Vero-TMPRSS2 cells, some minor differences in potency were noted in cells expressing hACE2.
Fig. 3. Neutralization of SARS-CoV-2 B.1.1.529 Omicron strain by mAbs in Vero-hACE2-TMPRSS2 cells.
a–f, Neutralization curves in Vero-hACE2-TMPRSS2 cells comparing the sensitivity of SARS-CoV-2 strains with the indicated mAbs (S309, COV2-2196, COV2-2130, REGN10933, REGN10987, LY-CoV555, LY-CoV016, CT-P59 and SARS2-38) with WA1/2020 D614G and B.1.1.529. Also shown are the neutralization curves for antibody cocktails (COV2-2196/COV2-2130, REGN10933/REGN10987 or LY-CoV555/LY-CoV016). For data with mAb combinations, the x axis represents the total concentration of mAb used. One representative experiment of three performed in technical duplicate is shown. Error bars indicate range of technical replicates. Data (% relative infection) are normalized to a no-mAb control. g, Summary of EC50 values (ng ml−1) of neutralization of SARS-CoV-2 viruses (WA1/2020 D614G and B.1.1.529) performed in Vero-hACE2-TMPRSS2 cells. Data are the geometric mean of three experiments. Blue shading: light, EC50 > 5,000 ng ml−1; dark, EC50 > 10,000 ng ml−1. h, Comparison of EC50 values by mAbs against WA1/2020 D614G and B.1.1.529. i, j, Neutralization curves in Vero-hACE2-TMPRSS2 cells comparing WA1/2020 D614G and B.1.1.529 infection in the presence of AZD1061, AZD8895 and the combination AZD7442. h, j, Three experiments; ****P < 0.0001; two-way ANOVA with Sidak’s post test. Each symbol represents neutralization data from an individual experiment. Bars indicate mean values. The dotted line indicates the upper limit of dosing of the assay.
Discussion
Our experiments show a marked loss of inhibitory activity by several of the most highly neutralizing mAbs that are in advanced clinical development or have EUA approval. We evaluated antibodies that correspond to monotherapy or combination therapy that have shown pre- and post-exposure success in clinical trials and in patients infected with historical SARS-CoV-2 isolates. Our results confirm in silico predictions of how amino acid changes in B.1.1.529 RBD might negatively affect neutralizing antibody interactions18,27. Moreover, they agree with preliminary studies showing that several clinically used antibodies lose neutralizing activity against B.1.1.529 spike-expressing recombinant lentiviral or vesicular stomatitis virus (VSV)-based pseudoviruses28–30. One difference is that our study with authentic B.1.1.529 showed only moderately reduced neutralization by antibodies corresponding to the AstraZeneca combination (COV2-2196 and COV2-2130); in contrast, another group reported escape of these mAbs using a VSV pseudovirus displaying a B.1.1.529 spike protein in Huh7 hepatoma cells29. Additional studies are needed to determine whether this disparity in results is due to the cell type, the virus (authentic versus pseudotype) or preparation and combination of antibody. To begin to address this issue, we obtained AZD1061, AZD8895 and the combination AZD7442 directly from the manufacturer and tested them for neutralization of WA1/2020 D614G and B.1.1529 in Vero-hACE2-TMPRSS2 cells. We observed relatively similar reductions in inhibitory activity compared to the preclinical COV2-2130 and COV2-2196 mAbs with 49-, 92- and 33-fold lower EC50 values against B.1.1.529 by AZD1061, AZD8895 and AZD7442, respectively (Fig. 3g,i–j).
Although the Regeneron (REGN10933 and REGN10987), Eli Lilly (LY-CoV555 and LV-CoV016) and Celltrion (CT-P59) antibodies or combinations showed an almost complete loss of neutralizing activity against B.1.1.529, in our assays with Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells the mAbs corresponding to the AstraZeneca combination (COV-2196 and COV-2130) or Vir Biotechnology (S309) products retained substantial inhibitory activity. Although these data suggest that some mAbs in clinical use might retain benefit, validation experiments in vivo7 are needed to support this conclusion and inform clinical decisions.
Given the loss of inhibitory activity against B.1.1.529 of many highly neutralizing anti-RBD mAbs in our study, it appears likely that serum polyclonal antibody responses generated after vaccination or natural infection also might lose substantial inhibitory activity against B.1.1.529, which could compromise protective immunity and explain a rise in symptomatic infections in vaccinated individuals31. Indeed, studies have reported approximately 25-fold to 40-fold reductions in serum neutralizing activity compared to historical D614G-containing strains from individuals immunized with the Pfizer BNT162b2 and AstraZeneca AZD1222 vaccines28,30,32,33.
We note several limitations of our study. (1) Our experiments focused on the effect of the extensive sequence changes in the B.1.1.529 spike protein on mAb neutralization in cell culture. Despite observing differences in neutralizing activity with certain mAbs, it remains to be determined how this finding translates into effects on clinical protection against B.1.1.529. (2) Although virus neutralization is a correlate of immune protection against SARS-CoV-2 (refs. 7,34,35), this measurement does not account for Fc effector functions if antibodies residually bind B.1.1.529 spike proteins on the virion or surface of infected cells. Fcγ receptor or complement protein engagement by spike-binding antibodies could confer substantial protection36–38. It should be noted that some antibodies have been engineered to have reduced Fc effector binding/function (e.g., the clinical antibodies AZD1061 and AZD8895). (3) We used the prevailing B.1.1.529 Omicron isolate that lacks an R346K mutation. Although only 8.3% of B.1.1.529 sequences in GISAID (accessed on 14 December 2021) have an R346K mutation, this substitution might further affect neutralization of some of the clinically used mAbs given that R346 is a contact residue for COV2-2130, REGN10987 and S309 (Fig. 1l). At least for S309, the R346K mutation did not affect neutralization of pseduoviruses displaying B.1.1.529 spike proteins30. Nonetheless, studies with infectious B.1.1.529 isolates with R346K mutations might be warranted if the substitution becomes more prevalent. (4) Our data are derived from experiments with Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells. Although these cells standardly are used to measure antibody neutralization of SARS-CoV-2 strains, primary cells targeted by SARS-CoV-2 in vivo can express unique sets of attachment and entry factors39, which could affect receptor and entry blockade by specific antibodies. Indeed, previous studies have reported that the cell line used can affect the potency of antibody neutralization against different SARS-CoV-2 variants6.
In summary, our cell-culture-based analysis of neutralizing mAb activity against an authentic infectious B.1.1.529 Omicron SARS-CoV-2 isolate suggests that several, but not all, existing therapeutic antibodies will lose protective benefit. Thus, the continued identification and use of broadly and potently neutralizing mAbs that target the most highly conserved residues on the SARS-CoV-2 spike likely is needed to prevent resistance against B.1.1.529 and future variants with highly mutated spike sequences.
Methods
Cells
Vero-TMPRSS2 (ref. 41) and Vero-hACE2-TMPRSS2 (ref. 6) cells were cultured at 37 °C in DMEM supplemented with 10% FBS, 10 mM HEPES pH 7.3 and 100 U ml−1 of penicillin–streptomycin. Vero-TMPRSS2 cells were supplemented with 5 μg ml−1 of blasticidin. Vero-hACE2-TMPRSS2 cells were supplemented with 10 µg ml−1 of puromycin. All cells routinely tested negative for mycoplasma using a PCR-based assay.
Viruses
The WA1/2020 recombinant strain with substitutions (D614G) was described previously42. The B.1.1.529 isolate (hCoV-19/USA/WI-WSLH-221686/2021) was obtained from an individual in Wisconsin as a midturbinate nasal swab and passaged once on Vero-TMPRSS2 cells43. All viruses were subjected to next-generation sequencing (GISAID: EPI_ISL_7263803) to confirm the stability of substitutions. All virus experiments were performed in an approved Biosafety Level 3 facility.
Monoclonal antibody purification
The mAbs used in this study (COV2-2196, COV2-2130, S309, REGN10933, REGN10987, LY-CoV555, LY-CoV016, CT-P59, SARS2-38, AZD1061, AZD8895 and AZD7442) have been described previously14,17,21,44–48. S309 is the parent of VIR-7831 (sotrovimab); the clinically used mAb is engineered for enhanced clinical developability, as reported previously23. COV2-2196 and COV2-2130 mAbs were produced after transient transfection using the Gibco ExpiCHO Expression System (Thermo Fisher Scientific) following the manufacturer’s protocol. Culture supernatants were purified using HiTrap MabSelect SuRe columns (Cytiva, formerly GE Healthcare Life Sciences) on an ÄKTA Pure chromatographer (GE Healthcare Life Sciences). Purified mAbs were buffer exchanged into PBS, concentrated using Amicon Ultra-4 50-kDa centrifugal filter units (Millipore Sigma) and stored at −80 °C until use. Purified mAbs were tested for endotoxin levels (found to be less than 30 endotoxin units (EU) per milligram IgG). Endotoxin testing was performed using the PTS201F cartridge (Charles River Laboratories), with a sensitivity range from 10 to 0.1 EU per milliliter, and an Endosafe Nexgen-MCS instrument (Charles River Laboratories). S309, REGN10933, REGN10987, LY-CoV016, LY-CoV555, CT-P59 and SARS2-38 mAb proteins were produced in CHOEXPI or EXPI293F cells and affinity purified using HiTrap Protein A columns (GE Healthcare, HiTrap mAb select Xtra no. 28-4082-61). Purified mAbs were suspended into 20 mM histidine, 8% sucrose pH 6.0 or PBS. The final products were sterilized by filtration through 0.22-μm filters and stored at 4 °C.
FRNT
Serial dilutions of mAbs were incubated with 102 focus-forming units of SARS-CoV-2 (WA1/2020 D614G or B.1.1.529) for 1 h at 37 °C. Antibody–virus complexes were added to Vero-TMPRSS2 or Vero-hACE2-TMPRSS2 cell monolayers in 96-well plates and incubated at 37 °C for 1 h. Subsequently, cells were overlaid with 1% (wt/vol) methylcellulose in MEM. Plates were harvested at 30 h (WA1/2020 D614G on Vero-TMPRSS2 cells), 70 h (B.1.1.529 on Vero-TMPRSS2 cells) or 24 h (both viruses on Vero-hACE2-TMPRSS2 cells) later by removal of overlays and fixation with 4% paraformaldehyde in PBS for 20 min at room temperature. A longer time of incubation was required for B.1.1.529-infected Vero-TMPRSS2 cells because the foci were smaller at the time point and difficult to quantitate. Plates with WA1/2020 D614G were washed and sequentially incubated with an oligoclonal pool (1 μg ml−1 of each) of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57 and SARS2-71 (ref. 49) anti-S antibodies. Plates with B.1.1.529 were additionally incubated with a pool of mAbs that cross-react with SARS-CoV-1 and bind a CR3022-competing epitope on the RBD21. All plates were subsequently stained with HRP-conjugated goat anti-mouse IgG (Sigma-Aldrich, A8924, 1:1,000) in PBS supplemented with 0.1% saponin and 0.1% BSA. SARS-CoV-2-infected cell foci were visualized using KPL TrueBlue peroxidase substrate and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies). Data (% relative infection) are normalized to a no-mAb control. Antibody dose–response curves were analyzed using non-linear regression analysis with a variable slope (GraphPad Software), and the EC50 was calculated.
Model of mAb-B.1.1.529 spike complexes
The spike model is a composite of data from PDB: 7C2L and PDB: 6W41. Models of mAb complexes were generated from their respective PDB files with the following accession codes: COV2-2196 (PDB: 7L7D); COV2-2130 (PDB: 7L7E); S309 (PDB: 6WPS); REGN10987 (PDB: 6XDG); REGN10933 (PDB: 6XDG); LY-CoV555 (PDB: 7KMG); LY-CoV016 (PDB: 7C01); CT-P59 (PDB: 7CM4); and SARS2-38 (PDB: 7MKM). Epitope footprints used in the multiple sequence alignment were determined using PISA interfacial analysis on the various mAb:RBD complexes50. Structural figures were generated using UCSF ChimeraX51.
Reagent availability
All reagents described in this paper are available through material transfer agreements. AZD8895 and AZD1061 may be obtained from AstraZeneca for non-commercial internal research purposes under material transfer agreements upon reasonable request.
Statistical analysis
The number of independent experiments and technical replicates used are indicated in the relevant figure legends. A two-way ANOVA with Sidak’s post test was used for comparisons of antibody potency between WA1/2020 D614G and B.1.1.59.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41591-021-01678-y.
Supplementary information
Acknowledgements
This study was supported by grants and contracts from the National Institues of Health (R01 AI157155 (to M.S.D. and J.E.C), U01 AI151810 (to M.S.D.), 75N93021C00014 (to Y.K.), HHSN272201700060C (to D.H.F.), 75N93019C00062 (D.H.F. and M.S.D.), and 75N93019C00051 (to M.S.D.)); the Defense Advanced Research Project Agency (HR0011-18-2-0001 (to J.E.C. and M.S.D)); the Japan Program for Infectious Diseases Research and Infrastructure (JP21wm0125002) from the Japan Agency for Medical Research and Development (to Y.K.); and the Dolly Parton COVID-19 Research Fund at Vanderbilt University Medical Center (to J.E.C.). We thank R. Nargi, R. Carnahan, T. Tan and L. Schimanski for assistance and generosity with mAb generation and purification and S. A. Turner from the Center for Pathogen Evolution at the University of Cambridge for evaluating B.1.1.529 sequences. AZD7442 (AZD8895 and AZD1061) was supplied by AstraZeneca under a material transfer agreement.
Source data
Raw data for neutralization curves
Raw data for neutralization curves
Author contributions
L.A.V. performed and analyzed neutralization assays. P.J.H. and L.A.V. propagated SARS-CoV-2 viruses. P.H. performed sequencing analysis. J.E.C., S.J.Z., L.P. and D.C. generated and provided mAbs. J.M.E. and D.H.F. performed structural analysis. J.E.C., D.H.F., Y.K. and M.S.D. obtained funding and supervised the research. L.A.V. and M.S.D. wrote the initial draft, with all other authors providing editorial comments.
Peer review
Peer review information
Nature Medicine thanks Julie Overbaugh, Barton Haynes and Sujan Shresta for their contribution to the peer review of this work. Editor recognition statement: João Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Data availability
All data supporting the findings of this study are available within the paper, in the Source Data and from the corresponding author upon reasonable request. There are no restrictions in obtaining access to primary data. Source data are provided with this paper.
Code availability
No code was used in the course of the data acquisition or analysis.
Competing interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology and Emergent BioSolutions. J.E.C. has served as a consultant for Luna Biologics and Merck Sharp & Dohme, is on the Scientific Advisory Board of Meissa Vaccines and is the founder of IDBiologics. The Crowe laboratory has received sponsored research agreements from Takeda Vaccines, AstraZeneca and IDBiologics. Vanderbilt University has applied for patents related to two antibodies discussed in this paper. L.A.P. and D.C. are employees of Vir Biotechnology and may hold stock shares in Vir Biotechnology. L.A.P. is a former employee and shareholder in Regeneron Pharmaceuticals. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41591-021-01678-y.
References
- 1.Sempowski GD, Saunders KO, Acharya P, Wiehe KJ, Haynes BF. Pandemic preparedness: developing vaccines and therapeutic antibodies for COVID-19. Cell. 2020;181:1458–1463. doi: 10.1016/j.cell.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wibmer CK, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 3.Wang, Z. et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature592, 616–622 (2021). [DOI] [PMC free article] [PubMed]
- 4.Tada, T. et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. Preprint at https://www.biorxiv.org/content/10.1101/2021.02.05.430003v1 (2021).
- 5.Wang, P. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature593, 130–135 (2021). [DOI] [PubMed]
- 6.Chen, R. E. et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med.27, 717–726 (2021). [DOI] [PMC free article] [PubMed]
- 7.Chen, R. E. et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature596, 103–108 (2021). [DOI] [PMC free article] [PubMed]
- 8.Callaway, E. & Ledford, H. How bad is Omicron? What scientists know so far. Nature600, 197–199 (2021). [DOI] [PubMed]
- 9.Torjesen, I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ375, n2943 (2021). [DOI] [PubMed]
- 10.Johnson, B. A. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature591, 293–299 (2021). [DOI] [PMC free article] [PubMed]
- 11.Chen, J., Wang, R., Gilby, N. B. & Wei, G. W. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. Preprint at https://arxiv.org/abs/2112.01318 (2021). [DOI] [PMC free article] [PubMed]
- 12.Barnes CO, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greaney AJ, et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto D, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 15.Gupta A, et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 16.Zost SJ, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum, A. et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science370, 1110–1115 (2020). [DOI] [PMC free article] [PubMed]
- 18.Starr, T. N., Greaney, A. J., Dingens, A. S. & Bloom, J. D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med.20, 100255 (2021). [DOI] [PMC free article] [PubMed]
- 19.Gottlieb RL, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim C, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanBlargan, L. A. et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity54, 2399–2416 (2021). [DOI] [PMC free article] [PubMed]
- 22.Case JB, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cathcart, A. L. et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. Preprint at https://www.biorxiv.org/content/10.1101/2021.03.09.434607v8 (2021).
- 24.Suryadevara N, et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golcuk, M., Yildiz, A. & Gur, M. The Omicron variant increases the interactions of SARS-CoV-2 spike glycoprotein with ACE2. Preprint at https://www.biorxiv.org/content/10.1101/2021.12.06.471377v1 (2021). [DOI] [PMC free article] [PubMed]
- 26.Lempp FA, et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 27.Ford, C. T., Machado, D. J. & Janies, D. A. Predictions of the SARS-CoV-2 Omicron variant (B.1.1.529) spike protein receptor-binding domain structure and neutralizing antibody interactions. Preprint at https://www.biorxiv.org/content/10.1101/2021.12.03.471024v3.full (2021).
- 28.Wilhelm, A. et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. Preprint at https://www.medrxiv.org/content/10.1101/2021.12.07.21267432v1 (2021).
- 29.Cao, Y. R. et al. B.1.1.529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes. Preprint at https://www.biorxiv.org/content/10.1101/2021.12.07.470392v1 (2021).
- 30.Cameroni, E. et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature10.1038/d41586-021-03825-4 (2021). [DOI] [PMC free article] [PubMed]
- 31.Callaway, E. Omicron likely to weaken COVID vaccine protection. Nature600, 367–368 (2021). [DOI] [PubMed]
- 32.Cele, S. et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. Preprint at https://www.medrxiv.org/content/10.1101/2021.12.08.21267417v1 (2021).
- 33.Dejnirattisai, W. et al. Reduced neutralisation of SARS-COV-2 Omicron-B.1.1.529 variant by post-immunisation serum. Preprint at https://www.medrxiv.org/content/10.1101/2021.12.10.21267534v1 (2021). [DOI] [PMC free article] [PubMed]
- 34.Kim, J. H., Marks, F. & Clemens, J. D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med.27, 205–211 (2021). [DOI] [PubMed]
- 35.Khoury DS, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 36.Schäfer, A. et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med.218, e20201993 (2021). [DOI] [PMC free article] [PubMed]
- 37.Zohar T, et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler ES, et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey AL, Diamond MS. A Crisp(r) new perspective on SARS-CoV-2 biology. Cell. 2021;184:15–17. doi: 10.1016/j.cell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 41.Zang, R., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol.5, eabc3582 (2020). [DOI] [PMC free article] [PubMed]
- 42.Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature592, 116–121 (2020). [DOI] [PMC free article] [PubMed]
- 43.Imai M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zost SJ, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tortorici MA, et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baum, A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science369, 1014–1018 (2020). [DOI] [PMC free article] [PubMed]
- 47.Jones BE, et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Trans. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi R, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 49.Liu Z, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper, in the Source Data and from the corresponding author upon reasonable request. There are no restrictions in obtaining access to primary data. Source data are provided with this paper.
No code was used in the course of the data acquisition or analysis.