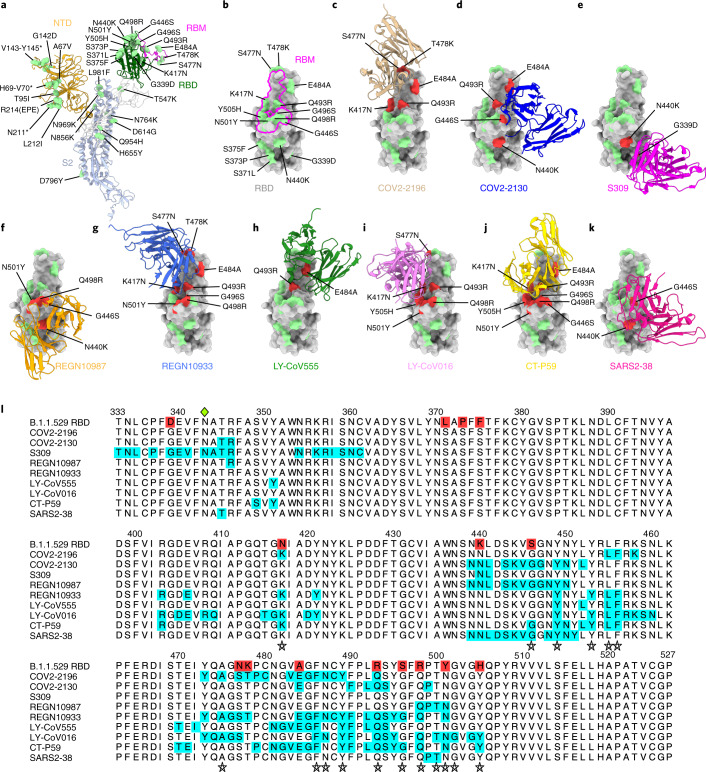
Fig. 1. Neutralizing mAb epitopes on B.1.1.529.
a, b, SARS-CoV-2 spike trimer (PDB: 7C2L and PDB: 6W41). One spike protomer is highlighted, showing the NTD in orange, RBD in green, RBM in magenta and S2 portion of the molecule in blue (a). Close-up view of the RBD with the RBM outlined in magenta (b). Amino acids that are changed in B.1.1.529 compared to WA1/2020 are indicated in light green (a, b), with the exception of N679K and P681H, which were not modeled in the structures used. c–k, SARS-CoV-2 RBD bound by EUA mAbs COV2-2196 (c, PDB: 7L7D); COV2-2130 (d, PDB: 7L7E); S309 (e, PDB: 6WPS); REGN10987 (f, PDB: 6XDG); REGN10933 (g, PDB: 6XDG); LY-CoV555 (h, PDB: 7KMG); LY-CoV016 (i, PDB: 7C01); CT-P59 (j PDB: 7CM4); and SARS2-38 (k, PDB: 7MKM). Residues mutated in the B.1.1.529 RBD and contained in these mAbsʼ respective epitopes are shaded red, whereas those outside the epitope are shaded green. l, Multiple sequence alignment showing the epitope footprints of each EUA mAb on the SARS-CoV-2 RBD highlighted in cyan. B.1.1.529 RBD is shown in the top row, with sequence changes relative to the wild-type RBD highlighted red. A green diamond indicates the location of the N-linked glycan at residue 343. Stars below the alignment indicate hACE2 contact residues on the SARS-CoV-2 RBD40.