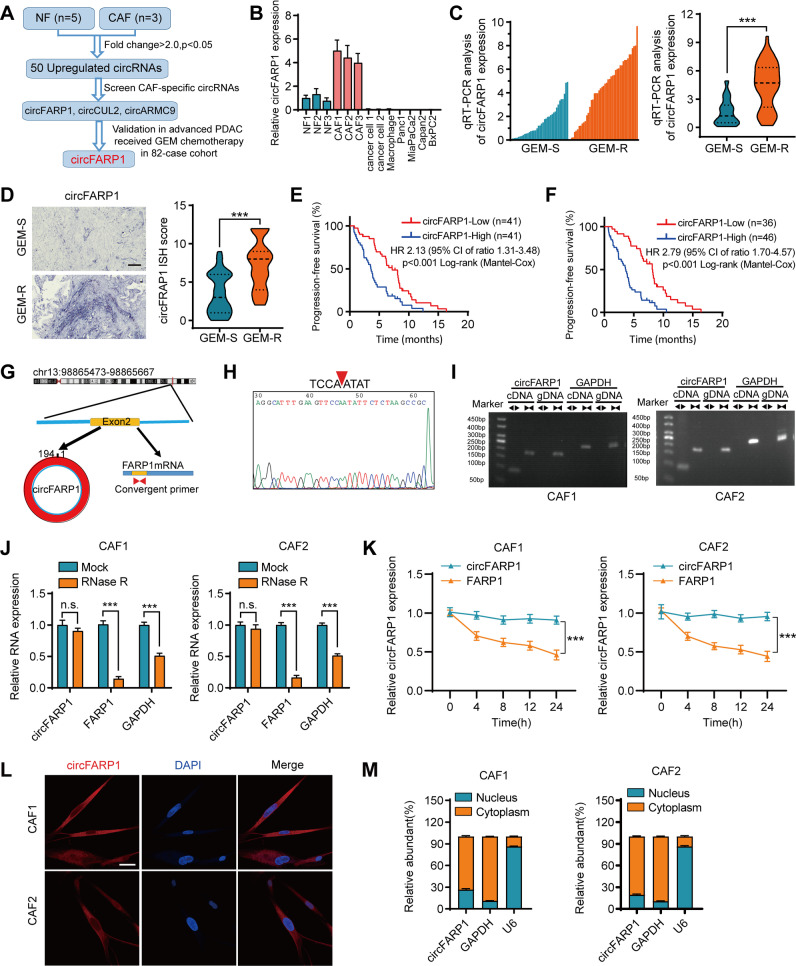
Fig. 1.
circFARP1 is overexpressed in CAFs and is associated with GEM chemoresistance and poor survival in advanced PDAC. A Schematic illustration of the identification of circFARP1 upregulated in CAFs. B qRT–PCR analysis of circFARP1 expression in NFs, CAFs, primary cancer cell, macrophages, and PDAC cell lines. C Quantification of circFARP1 expression by using qRT–PCR in GEM-S (n = 38) and GEM-R (n = 44) PDAC tissues. The left panel shows the plot of circFARP1 expression in each tissue. Right panel shows the expression as violin plots. D Representative images (left) and quantification (right) of circFARP1 by using ISH in GEM-S (n = 38) and GEM-R (n = 44) PDAC tissues. Scale bars, 50 μm. E-F Kaplan–Meier survival curves for advanced PDAC patients who received GEM-based chemotherapy with high or low circFARP1 expression evaluated by qRT–PCR (E) or ISH (F). A univariate Cox regression model was used to calculate the hazard ratio (HR). G Schematic illustration showing the genomic loci of circFARP1. CircFARP1 was generated by exons 2 of FARP1. H The back-splice junction of circFARP1 was identified by Sanger sequencing. I cDNA and gDNA of CAF1 and CAF2 were amplified with convergent and divergent primers. GAPDH was as the negative control. J PCR analysis of circFARP1, FARP1, and GAPDH expression in CAF1 and CAF2 cells treated with or without RNase R. n.s., not significant. K qRT–PCR analysis of circFARP1 and FARP1 mRNA in CAF1 and CAF2 cells treated with actinomycin D at the indicated time points. L Representative FISH images for circFARP1 in CAF1 and CAF2. Scale bars, 100 μm. M Subcellular fractionation assays of circFARP1 in CAF1 and CAF2. Data are expressed as the mean ± SD. ***p<0.001