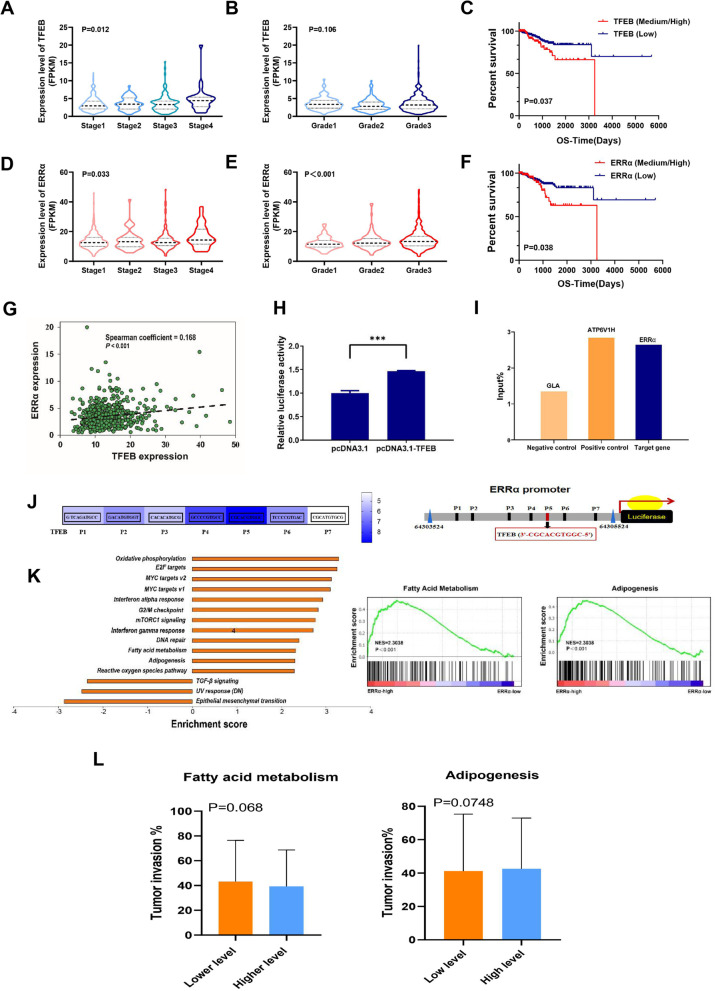
Fig. 1.
Bioinformatics analysis revealed that TFEB promotes ERRα transcription to participate in EC progression TCGA database (Sample size: Normal = 23; EC = 543) results are shown. (A) The expression of TFEB varies at different FIGO stages (B) and at different pathological grades. (C) The association of TFEB with OS in the patient/specimen quartiles is shown (Low: 1st quartile distribution; Median: 2nd-3rd quartile distribution; High: 4th quartile distribution). (D) The expression of ERRα varies at different FIGO stages (E) and at different pathological grades. (F) The association of ERRα with OS is shown. (G) The correlation between the expression levels of TFEB and ERRα in EC tissue. (H) ChIP analysis of the ERRα promoter occupancy in KLE cells is performed as described in the Materials and Methods section. TFEB is immunoprecipitated using an anti-Flag antibody, and DNA enrichment is performed using qPCR. The ATP6V1H promoter is used as a positive control and the GLA promoter is used as a negative control. (I) KLE cells are co-transfected with Flag-TFEB, ERRα promoter labeled with luciferase reporter, and Renilla luciferase control. 48 h after transfection, the cells are analyzed and the relative luciferase activity is measured and normalized to the Renilla luciferase control. (J) The putative ERRα-binding sites (ERREs), as predicted by the online program JASPAR (https://jaspar.genereg.net/analysis), are located in the TFEB (P1-P7) gene promoter regulatory regions. (K) KEGG pathway analysis (Ordinate: the KEGG signal path; abscissa: enrichment score). Results of GSEA in fatty acid metabolism and adipogenesis pathways. (L) The association of fatty acid metabolism and adipogenesis with tumor invasion is shown (Low: 1st- 2nd quartile distribution; High: 3rd-4th quartile distribution). Statistical tests: ANOVA (A-B, D-E), Kaplan–Meier estimator (C, F), Pearson correlation analysis (G), and Student’ s t-test (L). p < 0.05 suggests significantly different. TCGA: The Cancer Genome Atlas; FIGO: Federation International of Gynecology and Obstetrics; OS: Overall Survival; ChIP: Chromatin Immunoprecipitation; KEGG: Kyoto Encyclopaedia of Genes and Genomes; GSEA: Gene Set Enrichment Analysis.