Figure 1.
A conceptual illustration of the workflow to detect hub genes during cancer-stroma interaction
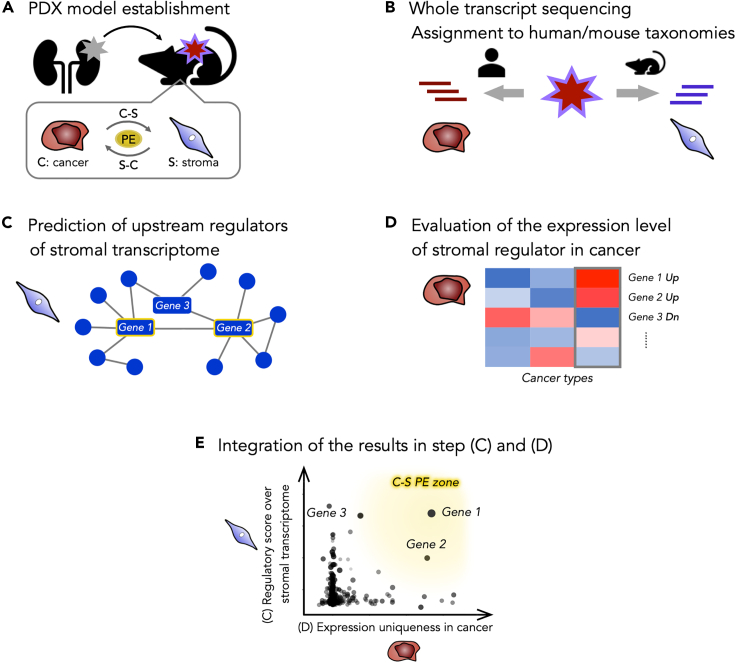
The pipeline consists of the following five steps.
(A): Patient-derived xenograft (PDX) model establishment. Here, 70 PDXs comprising nine different primary tumor types (COAD, NSCLC, EWS, PAAD, KIRC, STAC, GBM, GIST, and BRCA. See the Results section for abbreviation), were established. PE: Paracrine effectors.
(B): Transcript quantification and assignment to human/mouse taxonomies. Sequenced reads were quantified with reference to the combined transcriptome of human and mouse, then subsequently divided into the right taxonomy, representing cancer cell-derived (human) or stromal cell-derived (mouse) counts, respectively.
(C): Prediction of the upstream regulators of the stromal transcriptome. After a differential analysis conducted on the stroma of the PDX types of interest (e.g., the KIRC PDX model vs. the others), the upstream regulators over the detected differentially expressed (DE) genes were estimated using a directional pathway analysis (Ingenuity Upstream Regulator Analysis, IPA).
(D): Evaluation of the expression uniqueness of the predicted stromal regulator homologs in the complementary cancer cells. The differential analysis of the complementary cancer transcriptome (e.g., the KIRC PDX model versus the others) yielded a list of DE genes in cancer cells.
(E): Integration of the results of steps (C) and (D). The intersection of genes provided in steps (C) and (D) was depicted, with the scores of regulatory effects on the stromal transcriptome and those of the expression uniqueness in the cancer cells, in a scatterplot. The transcripts of the genes located in the right upper area (C-S PE zone) in the plot were considered more as paracrine effectors in the “cancer to stroma” interaction.