Figure 2.
The landscape of the transcriptome of patient-derived xenograft models
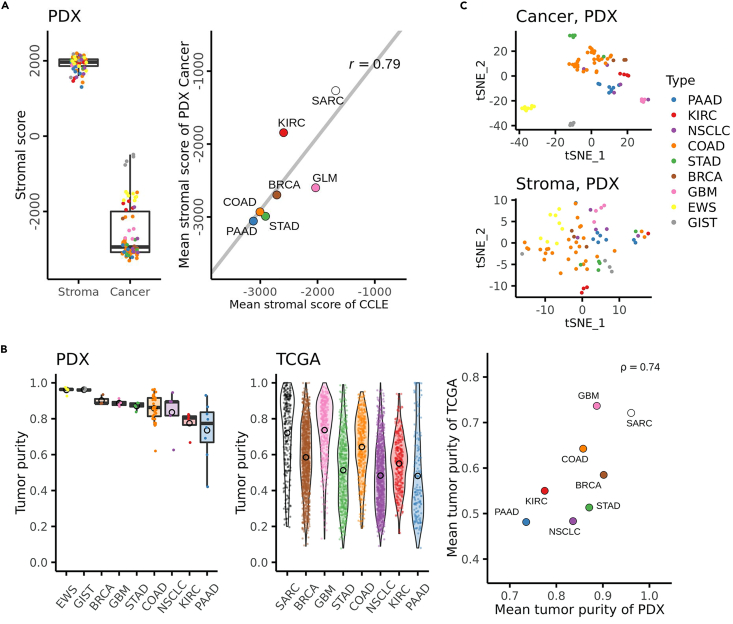
(A) The ESTIMATE stromal scores of stroma-assigned reads and cancer-assigned reads of PDXs (the left panel). The scatterplot showed mean stromal scores of PDX cancer components and corresponding cancer cell lines (CCLE) as references (the right panel). CCLE included PAAD (n = 41), COAD (n = 58), STAD (n = 41), BRCA (n = 56), KIRC (n = 25), GLM (65 gliomas corresponding to GBM in PDX), SARC (40 sarcomas corresponding to EWS/GIST in PDX). A linear regression line in gray and Pearson's correlation coefficient r are shown.
(B) Tumor purity of PDXs (the left panel) and non-xenografted cancers (TCGA) as a reference (the middle panel). The PDX tumor purity was defined as the ratio of the cancer gene-level count sum to the cancer plus stroma gene-level count sum. The TCGA tumor purity was computed using the ABSOLUTE algorithm. The scatterplot (the right panel) showed mean tumor purity values of PDX and TCGA, corresponding to small circles in black on the left and middle panels, respectively. The SARC in TCGA corresponds to the EWS and GIST in PDX. Spearman's correlation coefficient ρ is shown.
(C) The t-SNE plot of PDXs. Samples of the cancer component (the upper panel) or the stroma component (the lower panel) of the PDXs were plotted in t-SNE format, based on their transcriptome data, with a perplexity value of five. The colors of the dots were used to represent the PDX types as annotated.