Figure 3.
Tumor type-dependent characteristics of the PDX TMEs
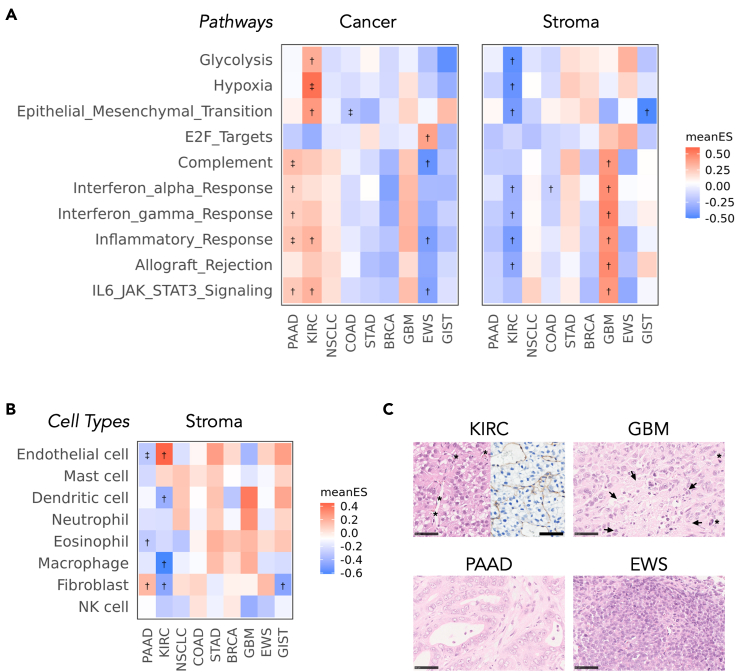
(A) Pathway analysis. The GSVA analysis was conducted for 40 of 50 hallmark pathways. The top 10 pathways measured to be statistically significant (q< 0.005, BH adjusted) using the one-way ANOVA test for PDX-types were shown.
(B) Cell type-specific gene set analysis. The GSVA analysis was conducted for gene sets specific to the eight major stromal cell types. The mean enrichment scores (meanES) of each PDX type, scaled to the sample-wise standard deviation (SD) and divided by the total SD, were shown by color scales. †; ‡(BH adjusted).
(C) Pathological images of the KIRC, PAAD, GBM, and EWS PDXs on hematoxylin and eosin (H&E) stain. The right half of the KIRC picture was stained using the anti-CD31 antibody. Asterisks (∗) denote intra-tumor vessels in the KIRC and mitotic tumor cells in the GBM. Arrows in the GBM show necrotic areas infiltrated by inflammatory cells. Scale bars represent 50 μm in length.